Real-time PCR in clinical microbiology: applications for routine laboratory testing
- PMID: 16418529
- PMCID: PMC1360278
- DOI: 10.1128/CMR.19.1.165-256.2006
Real-time PCR in clinical microbiology: applications for routine laboratory testing
Erratum in
- Clin Microbiol Rev. 2006 Jul;19(3):595
Abstract
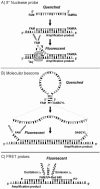
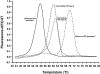
Real-time PCR has revolutionized the way clinical microbiology laboratories diagnose many human microbial infections. This testing method combines PCR chemistry with fluorescent probe detection of amplified product in the same reaction vessel. In general, both PCR and amplified product detection are completed in an hour or less, which is considerably faster than conventional PCR detection methods. Real-time PCR assays provide sensitivity and specificity equivalent to that of conventional PCR combined with Southern blot analysis, and since amplification and detection steps are performed in the same closed vessel, the risk of releasing amplified nucleic acids into the environment is negligible. The combination of excellent sensitivity and specificity, low contamination risk, and speed has made real-time PCR technology an appealing alternative to culture- or immunoassay-based testing methods for diagnosing many infectious diseases. This review focuses on the application of real-time PCR in the clinical microbiology laboratory.
Figures
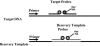
References
-
- Aberham, C., C. Pendl, P. Gross, G. Zerlauth, and M. Gessner. 2001. A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J. Virol. Methods 92:183-191. - PubMed
-
- Aberle, S. W., and E. Puchhammer-Stöckl. 2002. Diagnosis of herpesvirus infections of the central nervous system. J. Clin. Virol. 25(Suppl. 1):S79-85. - PubMed
-
- Aliyu, S. H., M. H. Aliyu, H. M. Salihu, S. Parmar, H. Jalal, and M. D. Curran. 2004. Rapid detection and quantitation of hepatitis B virus DNA by real-time PCR using a new fluorescent (FRET) detection system. J. Clin. Virol. 30:191-195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

