Coregulated human globin genes are frequently in spatial proximity when active
- PMID: 16418531
- PMCID: PMC2063548
- DOI: 10.1083/jcb.200507073
Coregulated human globin genes are frequently in spatial proximity when active
Abstract
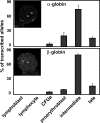
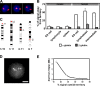
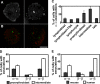
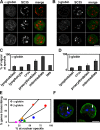
The organization of genes within the nucleus may influence transcription. We have analyzed the nuclear positioning of the coordinately regulated alpha- and beta-globin genes and show that the gene-dense chromatin surrounding the human alpha-globin genes is frequently decondensed, independent of transcription. Against this background, we show the frequent juxtaposition of active alpha- and beta-globin genes and of homologous alpha-globin loci that occurs at nuclear speckles and correlates with transcription. However, we did not see increased colocalization of signals, which would be expected with direct physical interaction. The same degree of proximity does not occur between human beta-globin genes or between murine globin genes, which are more constrained to their chromosome territories. Our findings suggest that the distribution of globin genes within erythroblast nuclei is the result of a self-organizing process, involving transcriptional status, diffusional ability of chromatin, and physical interactions with nuclear proteins, rather than a directed form of higher-order control.
Figures

References
-
- Boyle, S., S. Gilchrist, J. Bridger, N. Mahy, J. Ellis, and W. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. - PubMed
-
- Brown, J., K. Saracoglu, S. Uhrig, M. Speicher, R. Eils, and L. Kearney. 2001. Subtelomeric chromosome rearrangements are detected using an innovative 12-color FISH assay (M-TEL). Nat. Med. 7:497–501. - PubMed
-
- Brown, K., S. Guest, S. Smale, K. Hahm, M. Merkenschlager, and A. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. - PubMed

