A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks
- PMID: 16420670
- PMCID: PMC1414109
- DOI: 10.1186/gb-2005-6-13-r110
A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks
Abstract
Background: Transcription regulatory networks are composed of interactions between transcription factors and their target genes. Whereas unicellular networks have been studied extensively, metazoan transcription regulatory networks remain largely unexplored. Caenorhabditis elegans provides a powerful model to study such metazoan networks because its genome is completely sequenced and many functional genomic tools are available. While C. elegans gene predictions have undergone continuous refinement, this is not true for the annotation of functional transcription factors. The comprehensive identification of transcription factors is essential for the systematic mapping of transcription regulatory networks because it enables the creation of physical transcription factor resources that can be used in assays to map interactions between transcription factors and their target genes.
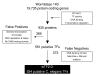
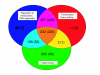
Results: By computational searches and extensive manual curation, we have identified a compendium of 934 transcription factor genes (referred to as wTF2.0). We find that manual curation drastically reduces the number of both false positive and false negative transcription factor predictions. We discuss how transcription factor splice variants and dimer formation may affect the total number of functional transcription factors. In contrast to mouse transcription factor genes, we find that C. elegans transcription factor genes do not undergo significantly more splicing than other genes. This difference may contribute to differences in organism complexity. We identify candidate redundant worm transcription factor genes and orthologous worm and human transcription factor pairs. Finally, we discuss how wTF2.0 can be used together with physical transcription factor clone resources to facilitate the systematic mapping of C. elegans transcription regulatory networks.
Conclusion: wTF2.0 provides a starting point to decipher the transcription regulatory networks that control metazoan development and function.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

