Discovery of biological networks from diverse functional genomic data
- PMID: 16420673
- PMCID: PMC1414113
- DOI: 10.1186/gb-2005-6-13-r114
Discovery of biological networks from diverse functional genomic data
Abstract
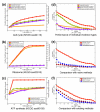
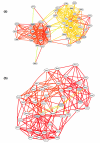
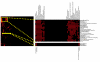
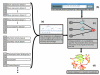
We have developed a general probabilistic system for query-based discovery of pathway-specific networks through integration of diverse genome-wide data. This framework was validated by accurately recovering known networks for 31 biological processes in Saccharomyces cerevisiae and experimentally verifying predictions for the process of chromosomal segregation. Our system, bioPIXIE, a public, comprehensive system for integration, analysis, and visualization of biological network predictions for S. cerevisiae, is freely accessible over the worldwide web.
Figures


References
-
- Jaimovich A, Elidan G, Margalit H, Friedman N. Towards an integrated protein-protein interaction network. In: Miyano S, Mesirov J, Kasif S, Istrail S, Pevzner P, Waterman M, editor. Research in Computational Molecular Biology: 9th Annual International Conference, RECOMB, Proceedings: May 14-18 2005, Cambridge, MA. Springer Verlag-GmbH; 2005. pp. 14–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

