A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities
- PMID: 16421248
- PMCID: PMC1415301
- DOI: 10.1091/mbc.e05-10-0956
A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities
Abstract
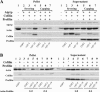
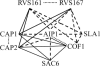
Actin interacting protein 1 (Aip1p) and cofilin cooperate to disassemble actin filaments in vitro and are thought to promote rapid turnover of actin networks in vivo. The precise method by which Aip1p participates in these activities has not been defined, although severing and barbed-end capping of actin filaments have been proposed. To better describe the mechanisms and biological consequences of Aip1p activities, we undertook an extensive mutagenesis of AIP1 aimed at disrupting and mapping Aip1p interactions. Site-directed mutagenesis suggested that Aip1p has two actin binding sites, the primary actin binding site lies on the edge of its N-terminal beta-propeller and a secondary actin binding site lies in a comparable location on its C-terminal beta-propeller. Random mutagenesis followed by screening for separation of function mutants led to the identification of several mutants specifically defective for interacting with cofilin but still able to interact with actin. These mutants suggested that cofilin binds across the cleft between the two propeller domains, leaving the actin binding sites exposed and flanking the cofilin binding site. Biochemical, genetic, and cell biological analyses confirmed that the actin binding- and cofilin binding-specific mutants are functionally defective, whereas the genetic analyses further suggested a role for Aip1p in an early, internalization step of endocytosis. A complementary, unbiased molecular modeling approach was used to derive putative structures for the Aip1p-cofilin complex, the most stable of which is completely consistent with the mutagenesis data. We theorize that Aip1p-severing activity may involve simultaneous binding to two actin subunits with cofilin wedged between the two actin binding sites of the N- and C-terminal propeller domains.
Figures




References
-
- Amatruda, J. F., Cannon, J. F., Tatchell, K., Hug, C., and Cooper, J. A. (1990). Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature 344, 352-354. - PubMed
-
- Amberg, D. C., Basart, E., and Botstein, D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28-35. - PubMed
-
- Amberg, D. C., Burke, D. J., and Strathern, J. N. (2005). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
-
- Baggett, J. J., and Wendland, B. (2001). Clathrin function in yeast endocytosis. Traffic 2, 297-302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

