COSMIC 2005
- PMID: 16421597
- PMCID: PMC2361125
- DOI: 10.1038/sj.bjc.6602928
COSMIC 2005
Abstract
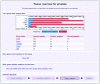
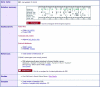
The Catalogue Of Somatic Mutations In Cancer (COSMIC) database and web site was developed to preserve somatic mutation data and share it with the community. Over the past 25 years, approximately 350 cancer genes have been identified, of which 311 are somatically mutated. COSMIC has been expanded and now holds data previously reported in the scientific literature for 28 known cancer genes. In addition, there is data from the systematic sequencing of 518 protein kinase genes. The total gene count in COSMIC stands at 538; 25 have a mutation frequency above 5% in one or more tumour type, no mutations were found in 333 genes and 180 are rarely mutated with frequencies <5% in any tumour set. The COSMIC web site has been expanded to give more views and summaries of the data and provide faster query routes and downloads. In addition, there is a new section describing mutations found through a screen of known cancer genes in 728 cancer cell lines including the NCI-60 set of cancer cell lines.
Figures
References
-
- Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE (2003) Mutational analysis of the tyrosine kinome in colorectal cancers. Science 300: 949. - PubMed
-
- Barnes DE, Lindahl T (2005) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Ann Rev Genet 38: 445–476 - PubMed
-
- Béroud C, Soussi T (2003) The UMD-p53 database: new mutations and analysis tools Hum Mutat. 21: 176–181 - PubMed
-
- Bignell G, Smith R, Hunter C, Stephens P, Davies H, Greenman C, Teague J, Butler A, Edkins S, Stevens C, O'meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Gillis AJ, Stoop HJ, van Gurp RJ, Oosterhuis JW, Looijenga LH, Futreal PA, Wooster R, Stratton MR (2005) Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer 45: 42–46 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

