Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1
- PMID: 16424388
- PMCID: PMC1895283
- DOI: 10.1182/blood-2005-11-4551
Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1
Abstract
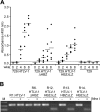
Natural antisense viral transcripts have been recognized in retroviruses, including human T-cell leukemia virus type 1 (HTLV-1), HIV-1, and feline immunodeficiency virus (FIV), and have been postulated to encode proteins important for the infection cycle and/or pathogenesis of the virus. The antisense strand of the HTLV-1 genome encodes HBZ, a novel nuclear basic region leucine zipper (b-ZIP) protein that in overexpression assays down-regulates Tax oncoprotein-induced viral transcription. Herein, we investigated the contribution of HBZ to HTLV-1-mediated immortalization of primary T lymphocytes in vitro and HTLV-1 infection in a rabbit animal model. HTLV-1 HBZ mutant viruses were generated and evaluated for viral gene expression, protein production, and immortalization capacity. Biologic properties of HBZ mutant viruses in vitro were indistinguishable from wild-type HTLV-1, providing the first direct evidence that HBZ is dispensable for viral replication and cellular immortalization. Rabbits inoculated with irradiated cells expressing HTLV-1 HBZ mutant viruses became persistently infected. However, these rabbits displayed a decreased antibody response to viral gene products and reduced proviral copies in peripheral blood mononuclear cells (PBMCs) as compared with wild-type HTLV-1-infected animals. Our findings indicated that HBZ was not required for in vitro cellular immortalization, but enhanced infectivity and persistence in inoculated rabbits. This study demonstrates that retroviruses use negative-strand-encoded proteins in the establishment of chronic viral infections.
Figures

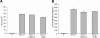
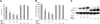

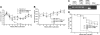
References
-
- Green PL, Chen ISY. Human T-cell leukemia virus types 1 and 2. In: Knipe DM, Howley P, Griffin D, Lamb R, Martin M, Straus S, eds. Virology. 4th ed. Philidelphia, PA: Lippincott Williams & Wilkins; 2001: 1941-1969.
-
- Collins ND, Newbound GC, Albrecht B, Beard J, Ratner L, Lairmore MD. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood. 1998;91: 4701-4707. - PubMed
-
- Wycuff DR, Marriott SJ. The HTLV-1 Tax oncoprotein: hypertasking at the molecular level. Front Biosci. 2005;10: 620-642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

