Unusual intron conservation near tissue-regulated exons found by splicing microarrays
- PMID: 16424921
- PMCID: PMC1331982
- DOI: 10.1371/journal.pcbi.0020004
Unusual intron conservation near tissue-regulated exons found by splicing microarrays
Abstract
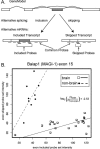
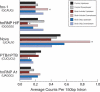
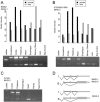
Alternative splicing contributes to both gene regulation and protein diversity. To discover broad relationships between regulation of alternative splicing and sequence conservation, we applied a systems approach, using oligonucleotide microarrays designed to capture splicing information across the mouse genome. In a set of 22 adult tissues, we observe differential expression of RNA containing at least two alternative splice junctions for about 40% of the 6,216 alternative events we could detect. Statistical comparisons identify 171 cassette exons whose inclusion or skipping is different in brain relative to other tissues and another 28 exons whose splicing is different in muscle. A subset of these exons is associated with unusual blocks of intron sequence whose conservation in vertebrates rivals that of protein-coding exons. By focusing on sets of exons with similar regulatory patterns, we have identified new sequence motifs implicated in brain and muscle splicing regulation. Of note is a motif that is strikingly similar to the branchpoint consensus but is located downstream of the 5' splice site of exons included in muscle. Analysis of three paralogous membrane-associated guanylate kinase genes reveals that each contains a paralogous tissue-regulated exon with a similar tissue inclusion pattern. While the intron sequences flanking these exons remain highly conserved among mammalian orthologs, the paralogous flanking intron sequences have diverged considerably, suggesting unusually complex evolution of the regulation of alternative splicing in multigene families.
Conflict of interest statement
Figures



References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
-
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. - PubMed
-
- Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19((Suppl 1)):i118–i121. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

