Foamy virus vector integration sites in normal human cells
- PMID: 16428288
- PMCID: PMC1360565
- DOI: 10.1073/pnas.0510046103
Foamy virus vector integration sites in normal human cells
Abstract
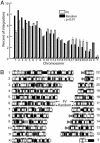
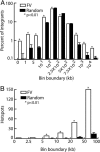
Foamy viruses (FVs) or spumaviruses are retroviruses that have been developed as vectors, but their integration patterns have not been described. We have performed a large-scale analysis of FV integration sites in unselected human fibroblasts (n = 1,008) and human CD34(+) hematopoietic cells (n = 1,821) by using a bacterial shuttle vector and a comparable analysis of lentiviral vector integration sites in CD34(+) cells (n = 1,331). FV vectors had a distinct integration profile relative to other types of retroviruses. They did not integrate preferentially within genes, despite a modest preference for integration near transcription start sites and a significant preference for CpG islands. The genomewide distribution of FV vector proviruses was nonrandom, with both clusters and gaps. Transcriptional profiling showed that gene expression had little influence on integration site selection. Our findings suggest that FV vectors may have desirable integration properties for gene therapy applications.
Figures



References
-
- Hacein-Bey-Abina, S., von Kalle, C., Schmidt, M., Le Deist, F., Wulffraat, N., McIntyre, E., Radford, I., Villeval, J. L., Fraser, C. C., Cavazzana-Calvo, M. & Fischer, A. (2003) N. Engl. J. Med. 348, 255-256. - PubMed
-
- Schroder, A. R., Shinn, P., Chen, H., Berry, C., Ecker, J. R. & Bushman, F. (2002) Cell 110, 521-529. - PubMed
-
- Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

