Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period
- PMID: 16428442
- PMCID: PMC1347041
- DOI: 10.1128/MCB.26.3.863-870.2006
Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period
Abstract
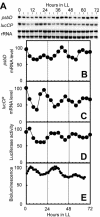
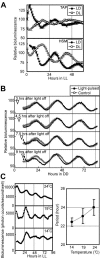
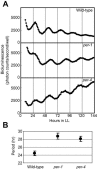
Chloroplast-encoded genes, like nucleus-encoded genes, exhibit circadian expression. How the circadian clock exerts its control over chloroplast gene expression, however, is poorly understood. To facilitate the study of chloroplast circadian gene expression, we developed a codon-optimized firefly luciferase gene for the chloroplast of Chlamydomonas reinhardtii as a real-time bioluminescence reporter and introduced it into the chloroplast genome. The bioluminescence of the reporter strain correlated well with the circadian expression pattern of the introduced gene and satisfied all three criteria for circadian rhythms. Moreover, the period of the rhythm was lengthened in per mutants, which are phototactic rhythm mutants carrying a long-period gene in their nuclear genome. These results demonstrate that chloroplast gene expression rhythm is a bona fide circadian rhythm and that the nucleus-encoded circadian oscillator determines the period length of the chloroplast rhythm. Our reporter strains can serve as a powerful tool not only for analysis of the circadian regulation mechanisms of chloroplast gene expression but also for a genetic approach to the molecular oscillator of the algal circadian clock.
Figures



References
-
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York, N.Y.
-
- Aoki, S., S. Kato, K. Ichikawa, and M. Shimizu. 2004. Circadian expression of the PpLhcb2 gene encoding a major light-harvesting chlorophyll a/b-binding protein in the moss Physcomitrella patens. Plant Cell Physiol. 45:68-76. - PubMed
-
- Bhattacharya, D., and L. Medlin. 1998. Algal phylogeny and the origin of land plants. Plant Physiol. 116:9-15.
-
- Brandes, C., J. D. Plautz, R. Stanewsky, C. F. Jamison, M. Straume, K. V. Wood, S. A. Kay, and J. C. Hall. 1996. Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron 16:687-692. - PubMed
-
- Bruce, V. G. 1970. The biological clock in Chlamydomonas reinhardi. J. Protozool. 17:328-334.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
