Mitochondrial transfer between cells can rescue aerobic respiration
- PMID: 16432190
- PMCID: PMC1345715
- DOI: 10.1073/pnas.0510511103
Mitochondrial transfer between cells can rescue aerobic respiration
Abstract
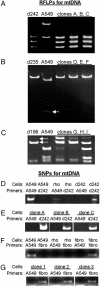
Current theory indicates that mitochondria were obtained 1.5 billion years ago from an ancient prokaryote. The mitochondria provided the capacity for aerobic respiration, the creation of the eukaryotic cell, and eventually complex multicellular organisms. Recent reports have found that mitochondria play essential roles in aging and determining lifespan. A variety of heritable and acquired diseases are linked to mitochondrial dysfunction. We report here that mitochondria are more dynamic than previously considered: mitochondria or mtDNA can move between cells. The active transfer from adult stem cells and somatic cells can rescue aerobic respiration in mammalian cells with nonfunctional mitochondria.
Figures



References
-
- Dyall, S. D., Brown, M. T. & Johnson, P. J. (2004) Science 304, 253–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

