Long-distance lateral diffusion of human Rad51 on double-stranded DNA
- PMID: 16432240
- PMCID: PMC1345706
- DOI: 10.1073/pnas.0508366103
Long-distance lateral diffusion of human Rad51 on double-stranded DNA
Abstract
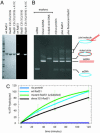
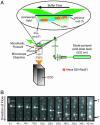
Rad51 is the primary eukaryotic recombinase responsible for initiating DNA strand exchange during homologous recombination. Although the subject of intense study for over a decade, many molecular details of the reactions promoted by Rad51 and related recombinases remain unknown. Using total internal reflection fluorescence microscopy, we directly visualized the behavior of individual Rad51 complexes on double-stranded DNA (dsDNA) molecules suspended in an extended configuration above a lipid bilayer. Here we show that complexes of Rad51 can bind to and slide freely along the helical axis of dsDNA. Sliding is bidirectional, does not require ATP hydrolysis, and displays properties consistent with a 1D random walk driven solely by thermal diffusion. The proteins move freely on the DNA for long periods of time; however, sliding terminates and the proteins become immobile upon encountering the free end of a linear dsDNA molecule. This study provides previously uncharacterized insights into the behaviors of human Rad51, which may apply to other members of the RecA-like family of recombinases.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

