The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p
- PMID: 16436507
- PMCID: PMC1415324
- DOI: 10.1091/mbc.e05-07-0668
The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p
Abstract
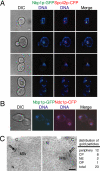
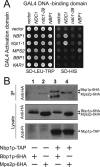
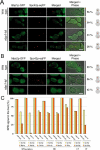
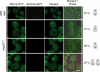
The spindle pole body (SPB) in Saccharomyces cerevisiae functions to nucleate and organize spindle microtubules, and it is embedded in the nuclear envelope throughout the yeast life cycle. However, the mechanism of membrane insertion of the SPB has not been elucidated. Ndc1p is an integral membrane protein that localizes to SPBs, and it is required for insertion of the SPB into the nuclear envelope during SPB duplication. To better understand the function of Ndc1p, we performed a dosage suppressor screen using the ndc1-39 temperature-sensitive allele. We identified an essential SPB component, Nbp1p. NBP1 shows genetic interactions with several SPB genes in addition to NDC1, and two-hybrid analysis revealed that Nbp1p binds to Ndc1p. Furthermore, Nbp1p is in the Mps2p-Bbp1p complex in the SPB. Immunoelectron microscopy confirmed that Nbp1p localizes to the SPB, suggesting a function at this location. Consistent with this hypothesis, nbp1-td (a degron allele) cells fail in SPB duplication upon depletion of Nbp1p. Importantly, these cells exhibit a "dead" SPB phenotype, similar to cells mutant in MPS2, NDC1, or BBP1. These results demonstrate that Nbp1p is a SPB component that acts in SPB duplication at the point of SPB insertion into the nuclear envelope.
Figures






References
-
- Adams, I. R., and Kilmartin, J. V. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10, 329-335. - PubMed
-
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1998). Current Protocols in Molecular Biology, New York: John Wiley & Sons.
-
- Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164-175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

