CMY-16, a novel acquired AmpC-type beta-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy
- PMID: 16436718
- PMCID: PMC1366893
- DOI: 10.1128/AAC.50.2.618-624.2006
CMY-16, a novel acquired AmpC-type beta-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy
Abstract
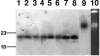
We report multifocal detection (four different cities in northern Italy) of Proteus mirabilis isolates resistant to both oxyimino- and 7-alpha-methoxy-cephalosporins and producing a novel acquired AmpC-like beta-lactamase. The enzyme, named CMY-16, is a variant of the CMY/LAT lineage, which differs from the closest homologues, CMY-4 and CMY-12, by a single amino acid substitution (A171S or N363S, respectively) and from CMY-2 by two substitutions (A171S and W221R). Expression of the cloned bla(CMY-16) gene in Escherichia coli decreased susceptibility to penicillins, cephalosporins, and aztreonam. Tazobactam was more effective than clavulanate at antagonizing the enzyme activity. Genotyping, by random amplification of polymorphic DNA and pulsed-field gel electrophoresis of genomic DNA digested with SfiI, showed that isolates were clonally related to each other, although not identical. The bla(CMY-16) gene was not transferable to E. coli by conjugation or transformation. In all isolates, it was chromosomally located and inserted in a conserved genetic environment. PCR mapping experiments revealed that the bla(CMY-16) was flanked by ISEcp1 and the blc gene, similar to other genes of this lineage from plasmids of Salmonella enterica, Klebsiella spp., and E. coli. Overall, these results revealed multifocal spreading of a CMY-16-producing P. mirabilis clone in northern Italy. This finding represents the first report of an acquired AmpC-like beta-lactamase in Proteus mirabilis from Italy and underscores the emergence of similar resistance determinants in the European setting.
Figures



References
-
- Bidet, P., C. Verdet, V. Gautier, E. Bingen, and G. Arlet. 2005. First description of DHA-1 ampC β-lactamase in Proteus mirabilis. Clin. Microbiol. Infect. 11:591-592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

