Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line
- PMID: 16440055
- PMCID: PMC1331978
- DOI: 10.1371/journal.pgen.0020002
Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line
Abstract
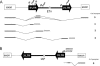
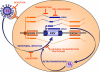
The inbred mouse is an invaluable model for human biology and disease. Nevertheless, when considering genetic mechanisms of variation and disease, it is important to appreciate the significant differences in the spectra of spontaneous mutations that distinguish these species. While insertions of transposable elements are responsible for only approximately 0.1% of de novo mutations in humans, the figure is 100-fold higher in the laboratory mouse. This striking difference is largely due to the ongoing activity of mouse endogenous retroviral elements. Here we briefly review mouse endogenous retroviruses (ERVs) and their influence on gene expression, analyze mechanisms of interaction between ERVs and the host cell, and summarize the variety of mutations caused by ERV insertions. The prevalence of mouse ERV activity indicates that the genome of the laboratory mouse is presently behind in the "arms race" against invasion.
Figures

References
-
- Brookfield JFY. The ecology of the genome—Mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–136. - PubMed
-
- Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. - PubMed
-
- Bartolome C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster . Mol Biol Evol. 2002;19:926–937. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

