Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency
- PMID: 16443695
- PMCID: PMC1400555
- DOI: 10.1104/pp.105.073825
Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency
Abstract
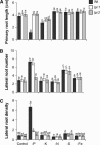
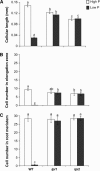
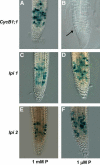
Low phosphorus (P) availability is one of the most limiting factors for plant productivity in many natural and agricultural ecosystems. Plants display a wide range of adaptive responses to cope with low P stress, which generally serve to enhance P availability in the soil and to increase its uptake by roots. In Arabidopsis (Arabidopsis thaliana), primary root growth inhibition and increased lateral root formation have been reported to occur in response to P limitation. To gain knowledge of the genetic mechanisms that regulate root architectural responses to P availability, we designed a screen for identifying Arabidopsis mutants that fail to arrest primary root growth when grown under low P conditions. Eleven low phosphorus insensitive (lpi) mutants that define at least four different complementation groups involved in primary root growth responses to P availability were identified. The lpi mutants do not show the typical determinate developmental program induced by P stress in the primary root. Other root developmental aspects of the low P rescue system, including increased root hair elongation and anthocyanin accumulation, remained unaltered in lpi mutants. In addition to the insensitivity of primary root growth inhibition, when subjected to P deprivation, lpi mutants show a reduced induction in the expression of several genes involved in the P starvation rescue system (PHOSPHATE TRANSPORTER 1 and 2, PURPLE ACID PHOSPHATASE 1, ACID PHOSPHATASE 5, and INDUCED BY PHOSPHATE STARVATION 1). Our results provide genetic support for the role of P as an important signal for postembryonic root development and root meristem maintenance and show a crosstalk in developmental and biochemical responses to P deprivation.
Figures

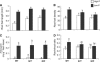

References
-
- Aida M, Beis D, Heidstra R, Williamsen V, Blilou I, Galinha C, Nussaume L, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 - PubMed
-
- Bariola PA, Howard CJ, Taylor CB, Verbug MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RSN1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 - PubMed
-
- Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorous availability. Plant Cell Environ 19: 529–538
-
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Chun-Ming L, Heidstra R, Sheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

