Subpopulation difference scanning: a strategy for exclusion mapping of susceptibility genes
- PMID: 16443857
- PMCID: PMC2564554
- DOI: 10.1136/jmg.2005.038414
Subpopulation difference scanning: a strategy for exclusion mapping of susceptibility genes
Abstract
Background: Association mapping is a common strategy for finding disease-related genes in complex disorders. Different association study designs exist, such as case-control studies or admixture mapping.
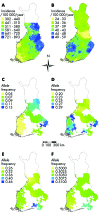
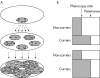
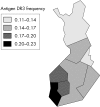
Methods: We propose a strategy, subpopulation difference scanning (SDS), to exclude large fractions of the genome as locations of genes for complex disorders. This strategy is applicable to genes explaining disease incidence differences within founder populations, for example, in cardiovascular diseases in Finland.
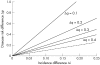
Results: The strategy consists of genotyping a set of markers from unrelated individuals sampled from subpopulations with differing disease incidence but otherwise as similar as possible. When comparing allele or haplotype frequencies between the subpopulations, the genomic areas with little difference can be excluded as possible locations for genes causing the difference in incidence, and other areas therefore targeted with case-control studies. As tests of this strategy, we use real and simulated data to show that under realistic assumptions of population history and disease risk parameters, the strategy saves efforts of sampling and genotyping and most efficiently detects genes of low risk--that is, those most difficult to find with other strategies.
Conclusion: In contrast to admixture mapping that uses the mixing of two different populations, the SDS strategy takes advantage of drift within highly related subpopulations.
Conflict of interest statement
Competing interests: there are no competing interests
Similar articles
-
Factors influencing the identification of major genes in a complex disease genome scan.Genet Epidemiol. 1997;14(6):933-8. doi: 10.1002/(SICI)1098-2272(1997)14:6<933::AID-GEPI62>3.0.CO;2-M. Genet Epidemiol. 1997. PMID: 9433603
-
Will admixture mapping work to find disease genes?Philos Trans R Soc Lond B Biol Sci. 2005 Aug 29;360(1460):1605-7. doi: 10.1098/rstb.2005.1691. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 16096110 Free PMC article. Review.
-
A genome-wide DNA microsatellite association screen to identify chromosomal regions harboring candidate genes in diabetic nephropathy.J Am Soc Nephrol. 2006 Mar;17(3):831-6. doi: 10.1681/ASN.2005050493. Epub 2006 Feb 8. J Am Soc Nephrol. 2006. PMID: 16467450
-
The genetic population structure of northern Sweden and its implications for mapping genetic diseases.Hereditas. 2007 Nov;144(5):171-80. doi: 10.1111/j.2007.0018-0661.02007.x. Hereditas. 2007. PMID: 18031350
-
Finding genes influencing susceptibility to complex diseases in the post-genome era.Am J Pharmacogenomics. 2001;1(3):203-21. doi: 10.2165/00129785-200101030-00005. Am J Pharmacogenomics. 2001. PMID: 12083968 Review.
References
-
- Risch N J. Searching for genetic determinants in the new millennium. Nature 2000405847–856. - PubMed
-
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 200333(suppl)228–237. - PubMed
-
- Nevanlinna H R. The Finnish population structure. A genetic and genealogical study. Hereditas 197271195–236. - PubMed
-
- Kere J. Human population genetics: lessons from Finland. Annu Rev Genomics Hum Genet 20012103–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials