Asynchronous nuclear division cycles in multinucleated cells
- PMID: 16449188
- PMCID: PMC2063645
- DOI: 10.1083/jcb.200507003
Asynchronous nuclear division cycles in multinucleated cells
Abstract
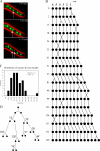
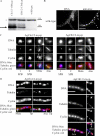
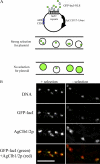
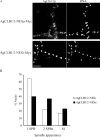
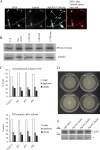
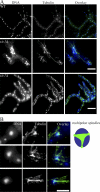
Synchronous mitosis is common in multinucleated cells. We analyzed a unique asynchronous nuclear division cycle in a multinucleated filamentous fungus, Ashbya gossypii. Nuclear pedigree analysis and observation of GFP-labeled spindle pole bodies demonstrated that neighboring nuclei in A. gossypii cells are in different cell cycle stages despite close physical proximity. Neighboring nuclei did not differ significantly in their patterns of cyclin protein localization such that both G1 and mitotic cyclins were present regardless of cell cycle stage, suggesting that the complete destruction of cyclins is not occurring in this system. Indeed, the expression of mitotic cyclin lacking NH(2)-terminal destruction box sequences did not block cell cycle progression. Cells lacking AgSic1p, a predicted cyclin-dependent kinase (CDK) inhibitor, however, showed aberrant multipolar spindles and fragmented nuclei that are indicative of flawed mitoses. We hypothesize that the continuous cytoplasm in these cells promoted the evolution of a nuclear division cycle in which CDK inhibitors primarily control CDK activity rather than oscillating mitotic cyclin proteins.
Figures



Similar articles
-
Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells.Eukaryot Cell. 2007 Mar;6(3):473-86. doi: 10.1128/EC.00273-06. Epub 2006 Nov 22. Eukaryot Cell. 2007. PMID: 17122387 Free PMC article.
-
Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit.J Cell Biol. 2006 Jan 30;172(3):335-46. doi: 10.1083/jcb.200507162. J Cell Biol. 2006. PMID: 16449187 Free PMC article.
-
Cytoplasmic Clb2 is required for timely inactivation of the mitotic inhibitor Swe1 and normal bud morphogenesis in Saccharomyces cerevisiae.Curr Genet. 2007 Jan;51(1):1-18. doi: 10.1007/s00294-006-0102-1. Epub 2006 Oct 11. Curr Genet. 2007. PMID: 17033818
-
Nuclear anarchy: asynchronous mitosis in multinucleated fungal hyphae.Curr Opin Microbiol. 2006 Dec;9(6):547-52. doi: 10.1016/j.mib.2006.09.002. Epub 2006 Oct 11. Curr Opin Microbiol. 2006. PMID: 17045513 Review.
-
Cyclin/Cdk complexes: their involvement in cell cycle progression and mitotic division.Protoplasma. 2001;216(3-4):119-42. doi: 10.1007/BF02673865. Protoplasma. 2001. PMID: 11732181 Review.
Cited by
-
Nuclear dynamics during germination, conidiation, and hyphal fusion of Fusarium oxysporum.Eukaryot Cell. 2010 Aug;9(8):1216-24. doi: 10.1128/EC.00040-10. Epub 2010 Jun 11. Eukaryot Cell. 2010. PMID: 20543061 Free PMC article.
-
Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells.Eukaryot Cell. 2007 Mar;6(3):473-86. doi: 10.1128/EC.00273-06. Epub 2006 Nov 22. Eukaryot Cell. 2007. PMID: 17122387 Free PMC article.
-
A conserved G₁ regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm.Cell Cycle. 2010 Sep 15;9(18):3771-9. doi: 10.4161/cc.9.18.12999. Epub 2010 Sep 13. Cell Cycle. 2010. PMID: 20930528 Free PMC article.
-
Of bars and rings: Hof1-dependent cytokinesis in multiseptated hyphae of Ashbya gossypii.Mol Cell Biol. 2009 Feb;29(3):771-83. doi: 10.1128/MCB.01150-08. Epub 2008 Nov 24. Mol Cell Biol. 2009. PMID: 19029253 Free PMC article.
-
Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation.PLoS Biol. 2018 Mar 29;16(3):e2003782. doi: 10.1371/journal.pbio.2003782. eCollection 2018 Mar. PLoS Biol. 2018. PMID: 29596410 Free PMC article.
References
-
- Alberti-Segui, C., F. Dietrich, R. Altmann-Johl, D. Hoepfner, and P. Philippsen. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975–986. - PubMed
-
- Altmann-Johl, R., and P. Philippsen. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69–80. - PubMed
-
- Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563–4575. - PubMed
-
- Belmont, A.S., and A.F. Straight. 1998. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 8:121–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

