Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels
- PMID: 16449388
- PMCID: PMC1413637
- DOI: 10.1073/pnas.0508483103
Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels
Abstract
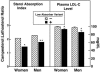
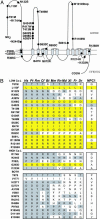
An approach to understand quantitative traits was recently proposed based on the finding that nonsynonymous (NS) sequence variants in certain genes are preferentially enriched at one extreme of the population distribution. The NS variants, although individually rare, are cumulatively frequent and influence quantitative traits, such as plasma lipoprotein levels. Here, we use the NS variant technique to demonstrate that genetic variation in NPC1L1 contributes to variability in cholesterol absorption and plasma levels of low-density lipoproteins (LDLs). The ratio of plasma campesterol (a plant sterol) to lathosterol (a cholesterol precursor) was used to estimate relative cholesterol absorption in a population-based study. Nonsynonymous sequence variations in NPC1L1 were five times more common in low absorbers (n = 26 of 256) than in high absorbers (n = 5 of 256) (P < 0.001). The rare variants identified in low absorbers were found in 6% of 1,832 African-Americans and were associated with lower plasma levels of LDL cholesterol (LDL-C) (96 +/- 36 mg/dl vs. 105 +/- 36 mg/dl; P = 0.005). These data, together with prior findings, reveal a genetic architecture for LDL-C levels that does not conform to current models for quantitative traits and indicate that a significant fraction of genetic variance in LDL-C is due to multiple alleles with modest effects that are present at low frequencies in the population.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Bosner M. S., Lange L. G., Stenson W. F., Ostlund R. E., Jr J. Lipid Res. 1999;40:302–308. - PubMed
-
- Ezzet F., Wexler D., Statkevich P., Kosoglou T., Patrick J., Lipka L., Mellars L., Veltri E., Batra V. J. Clin. Pharmacol. 2001;41:943–949. - PubMed
-
- Gylling H., Miettinen T. A. J. Lipid Res. 2002;43:1472–1476. - PubMed
-
- Altmann S. W., Davis H. R., Jr, Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. Science. 2004;303:1201–1204. - PubMed
-
- Davis H. R., Jr, Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. J. Biol. Chem. 2004;279:33586–33592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

