Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis
- PMID: 16449634
- PMCID: PMC1367182
- DOI: 10.1128/MCB.26.4.1183-1194.2006
Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis
Abstract
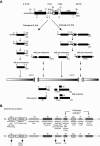
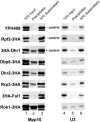
The 17 putative RNA helicases required for pre-rRNA processing are predicted to play a crucial role in ribosome biogenesis by driving structural rearrangements within preribosomes. To better understand the function of these proteins, we have generated a battery of mutations in five putative RNA helicases involved in 18S rRNA synthesis and analyzed their effects on cell growth and pre-rRNA processing. Our results define functionally important residues within conserved motifs and demonstrate that lethal mutations in predicted ATP binding-hydrolysis motifs often confer a dominant negative phenotype in vivo when overexpressed in a wild-type background. We show that dominant negative mutants delay processing of the 35S pre-rRNA and cause accumulation of pre-rRNA species that normally have low steady-state levels. Our combined results establish that not all conserved domains function identically in each protein, suggesting that the RNA helicases may have distinct biochemical properties and diverse roles in ribosome biogenesis.
Figures
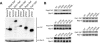




Similar articles
-
DExD/H-box RNA helicases in ribosome biogenesis.RNA Biol. 2013 Jan;10(1):4-18. doi: 10.4161/rna.21879. Epub 2012 Aug 24. RNA Biol. 2013. PMID: 22922795 Free PMC article. Review.
-
Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis.Mol Cell Biol. 2006 Feb;26(4):1195-208. doi: 10.1128/MCB.26.4.1195-1208.2006. Mol Cell Biol. 2006. PMID: 16449635 Free PMC article.
-
Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.Mol Microbiol. 2004 Apr;52(1):141-58. doi: 10.1111/j.1365-2958.2003.03973.x. Mol Microbiol. 2004. PMID: 15049817
-
Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis.Nucleic Acids Res. 2001 Mar 1;29(5):1144-55. doi: 10.1093/nar/29.5.1144. Nucleic Acids Res. 2001. PMID: 11222764 Free PMC article.
-
Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives.Biochim Biophys Acta. 2013 Aug;1829(8):775-90. doi: 10.1016/j.bbagrm.2013.01.007. Epub 2013 Jan 26. Biochim Biophys Acta. 2013. PMID: 23357782 Review.
Cited by
-
The RNA helicase Dbp10 coordinates assembly factor association with PTC maturation during ribosome biogenesis.Nucleic Acids Res. 2024 Feb 28;52(4):1975-1987. doi: 10.1093/nar/gkad1206. Nucleic Acids Res. 2024. PMID: 38113283 Free PMC article.
-
Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis.RNA. 2019 Jun;25(6):685-701. doi: 10.1261/rna.069609.118. Epub 2019 Mar 25. RNA. 2019. PMID: 30910870 Free PMC article.
-
DExD/H-box RNA helicases in ribosome biogenesis.RNA Biol. 2013 Jan;10(1):4-18. doi: 10.4161/rna.21879. Epub 2012 Aug 24. RNA Biol. 2013. PMID: 22922795 Free PMC article. Review.
-
The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot.PLoS Biol. 2015 Feb 24;13(2):e1002083. doi: 10.1371/journal.pbio.1002083. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25710520 Free PMC article.
-
The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis.Nucleic Acids Res. 2006 Jun 13;34(10):3189-99. doi: 10.1093/nar/gkl419. Print 2006. Nucleic Acids Res. 2006. PMID: 16772403 Free PMC article.
References
-
- Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
