Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus
- PMID: 16449639
- PMCID: PMC1367202
- DOI: 10.1128/MCB.26.4.1245-1258.2006
Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus
Abstract
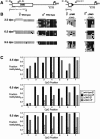
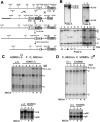
The differentially methylated domain (DMD) of the mouse H19 gene is a methylation-sensitive insulator that blocks access of the Igf2 gene to shared enhancers on the maternal allele and inactivates H19 expression on the methylated paternal allele. By analyzing H19 DMD deletion alleles H19DeltaDMD and H19Delta3.8kb-5'H19 in pre- and postimplantation embryos, we show that the DMD exhibits positive transcriptional activity and is required for H19 expression in blastocysts and full activation of H19 during subsequent development. We also show that the DMD is required to establish Igf2 imprinting by blocking access to shared enhancers when Igf2 monoallelic expression is initiated in postimplantation embryos and that the single remaining CTCF site of the H19DeltaDMD allele is unable to provide this function. Furthermore, our data demonstrate that sequence outside of the DMD can attract some paternal-allele-specific CpG methylation 5' of H19 in preimplantation embryos, although this methylation is not maintained during postimplantation in the absence of the DMD. Finally, we report a conditional allele floxing the 1.6-kb sequence deleted from the H19DeltaDMD allele and demonstrate that the DMD is required to maintain repression of the maternal Igf2 allele and the full activity of the paternal Igf2 allele in neonatal liver.
Figures




Similar articles
-
Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting.Mol Cell Biol. 2002 Apr;22(8):2450-62. doi: 10.1128/MCB.22.8.2450-2462.2002. Mol Cell Biol. 2002. PMID: 11909940 Free PMC article.
-
CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus.Hum Mol Genet. 2006 Oct 1;15(19):2945-54. doi: 10.1093/hmg/ddl237. Epub 2006 Aug 23. Hum Mol Genet. 2006. PMID: 16928784
-
Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3.PLoS Genet. 2007 Aug;3(8):e137. doi: 10.1371/journal.pgen.0030137. Epub 2007 Jun 29. PLoS Genet. 2007. PMID: 17708683 Free PMC article.
-
Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation.J Biochem. 2000 May;127(5):711-5. doi: 10.1093/oxfordjournals.jbchem.a022661. J Biochem. 2000. PMID: 10788777 Review.
-
Igf2/H19 imprinting control region (ICR): an insulator or a position-dependent silencer?ScientificWorldJournal. 2001 Jun 5;1:218-24. doi: 10.1100/tsw.2001.50. ScientificWorldJournal. 2001. PMID: 12806090 Free PMC article. Review.
Cited by
-
Progression to adrenocortical tumorigenesis in mice and humans through insulin-like growth factor 2 and β-catenin.Am J Pathol. 2012 Sep;181(3):1017-33. doi: 10.1016/j.ajpath.2012.05.026. Epub 2012 Jul 15. Am J Pathol. 2012. PMID: 22800756 Free PMC article.
-
Domain-specific response of imprinted genes to reduced DNMT1.Mol Cell Biol. 2010 Aug;30(16):3916-28. doi: 10.1128/MCB.01278-09. Epub 2010 Jun 14. Mol Cell Biol. 2010. PMID: 20547750 Free PMC article.
-
Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer.PLoS One. 2008 Mar 26;3(3):e1849. doi: 10.1371/journal.pone.0001849. PLoS One. 2008. PMID: 18365005 Free PMC article.
-
Tissue-specific and mosaic imprinting defects underlie opposite congenital growth disorders in mice.PLoS Genet. 2018 Feb 22;14(2):e1007243. doi: 10.1371/journal.pgen.1007243. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29470501 Free PMC article.
-
Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant.Genes Dev. 2016 Mar 1;30(5):567-78. doi: 10.1101/gad.275958.115. Genes Dev. 2016. PMID: 26944681 Free PMC article.
References
-
- Auffray, C., and F. Rougeon. 1980. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur. J. Biochem. 107:303-314. - PubMed
-
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
-
- Cho, H., J. Mu, J. K. Kim, J. L. Thorvaldsen, Q. Chu, E. B. Crenshaw III, K. H. Kaestner, M. S. Bartolomei, G. I. Shulman, and M. J. Birnbaum. 2001. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292:1728-1731. - PubMed
-
- Cranston, M. J., T. L. Spinka, D. A. Elson, and M. S. Bartolomei. 2001. Elucidation of the minimal sequence required to imprint H19 transgenes. Genomics 73:98-107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
