Chromatin decondensation and nuclear reprogramming by nucleoplasmin
- PMID: 16449640
- PMCID: PMC1367201
- DOI: 10.1128/MCB.26.4.1259-1271.2006
Chromatin decondensation and nuclear reprogramming by nucleoplasmin
Erratum in
- Mol Cell Biol. 2007 Sep;27(18):6580
Abstract
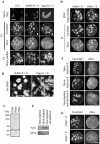
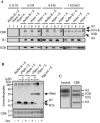
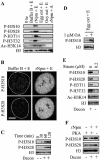
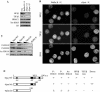
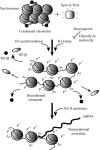
Somatic cell nuclear cloning has repeatedly demonstrated striking reversibility of epigenetic regulation of cell differentiation. Upon injection into eggs, the donor nuclei exhibit global chromatin decondensation, which might contribute to reprogramming the nuclei by derepressing dormant genes. Decondensation of sperm chromatin in eggs is explained by the replacement of sperm-specific histone variants with egg-type histones by the egg protein nucleoplasmin (Npm). However, little is known about the mechanisms of chromatin decondensation in somatic nuclei that do not contain condensation-specific histone variants. Here we found that Npm could widely decondense chromatin in undifferentiated mouse cells without overt histone exchanges but with specific epigenetic modifications that are relevant to open chromatin structure. These modifications included nucleus-wide multiple histone H3 phosphorylation, acetylation of Lys 14 in histone H3, and release of heterochromatin proteins HP1beta and TIF1beta from the nuclei. The protein kinase inhibitor staurosporine inhibited chromatin decondensation and these epigenetic modifications with the exception of H3 acetylation, potentially linking these chromatin events. At the functional level, Npm pretreatment of mouse nuclei facilitated activation of four oocyte-specific genes from the nuclei injected into Xenopus laevis oocytes. Future molecular elucidation of chromatin decondensation by Npm will significantly contribute to our understanding of the plasticity of cell differentiation.
Figures

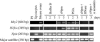
Similar articles
-
Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H1(0) from chromatin and the acquisition of transcriptional competence.EMBO J. 1996 Nov 1;15(21):5897-906. EMBO J. 1996. PMID: 8918467 Free PMC article.
-
Chromatin remodeling in somatic cells injected into mature pig oocytes.Reproduction. 2006 Jun;131(6):1037-49. doi: 10.1530/rep.1.00897. Reproduction. 2006. PMID: 16735543
-
Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization.J Biol Chem. 1996 Mar 29;271(13):7253-6. doi: 10.1074/jbc.271.13.7253. J Biol Chem. 1996. PMID: 8631735
-
The role of nucleoplasmin in chromatin assembly and disassembly.Philos Trans R Soc Lond B Biol Sci. 1993 Mar 29;339(1289):263-9; discussion 268-9. doi: 10.1098/rstb.1993.0024. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8098530 Review.
-
H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos.Nucleus. 2014 Sep-Oct;5(5):369-75. doi: 10.4161/nucl.36231. Nucleus. 2014. PMID: 25482190 Review.
Cited by
-
RE1-silencing Transcription Factor (REST) Is Required for Nuclear Reprogramming by Inhibiting Transforming Growth Factor β Signaling Pathway.J Biol Chem. 2016 Dec 30;291(53):27334-27342. doi: 10.1074/jbc.M116.743849. Epub 2016 Nov 7. J Biol Chem. 2016. PMID: 27821591 Free PMC article.
-
Reprogramming and development in nuclear transfer embryos and in interspecific systems.Curr Opin Genet Dev. 2012 Oct;22(5):450-8. doi: 10.1016/j.gde.2012.09.002. Epub 2012 Oct 11. Curr Opin Genet Dev. 2012. PMID: 23062626 Free PMC article. Review.
-
Dimerization of MORC2 through its C-terminal coiled-coil domain enhances chromatin dynamics and promotes DNA repair.Cell Commun Signal. 2019 Dec 3;17(1):160. doi: 10.1186/s12964-019-0477-5. Cell Commun Signal. 2019. PMID: 31796101 Free PMC article.
-
Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process?Nat Rev Mol Cell Biol. 2011 Jun 23;12(7):453-9. doi: 10.1038/nrm3140. Nat Rev Mol Cell Biol. 2011. PMID: 21697902 Free PMC article. Review.
-
Fatal familial insomnia: mitochondrial and protein synthesis machinery decline in the mediodorsal thalamus.Brain Pathol. 2017 Jan;27(1):95-106. doi: 10.1111/bpa.12408. Epub 2016 Aug 2. Brain Pathol. 2017. PMID: 27338255 Free PMC article.
References
-
- Ajiro, K., K. Yoda, K. Utsumi, and Y. Nishikawa. 1996. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 271:13197-13201. - PubMed
-
- Arnan, C., N. Saperas, C. Prieto, M. Chiva, and J. Ausio. 2003. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 278:31319-31324. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Banuelos, S., A. Hierro, J. M. Arizmendi, G. Montoya, A. Prado, and A. Muga. 2003. Activation mechanism of the nuclear chaperone nucleoplasmin: role of the core domain. J. Mol. Biol. 334:585-593. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
