Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron
- PMID: 16449646
- PMCID: PMC1367187
- DOI: 10.1128/MCB.26.4.1333-1346.2006
Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron
Abstract
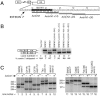
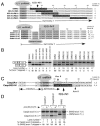
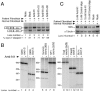
Humans have two nearly identical copies of the Survival Motor Neuron (SMN) gene, SMN1 and SMN2. In spinal muscular atrophy (SMA), SMN2 is not able to compensate for the loss of SMN1 due to exclusion of exon 7. Here we describe a novel inhibitory element located immediately downstream of the 5' splice site in intron 7. We call this element intronic splicing silencer N1 (ISS-N1). Deletion of ISS-N1 promoted exon 7 inclusion in mRNAs derived from the SMN2 minigene. Underlining the dominant role of ISS-N1 in exon 7 skipping, abrogation of a number of positive cis elements was tolerated when ISS-N1 was deleted. Confirming the silencer function of ISS-N1, an antisense oligonucleotide against ISS-N1 restored exon 7 inclusion in mRNAs derived from the SMN2 minigene or from endogenous SMN2. Consistently, this oligonucleotide increased the levels of SMN protein in SMA patient-derived cells that carry only the SMN2 gene. Our findings underscore for the first time the profound impact of an evolutionarily nonconserved intronic element on SMN2 exon 7 splicing. Considering that oligonucleotides annealing to intronic sequences do not interfere with exon-junction complex formation or mRNA transport and translation, ISS-N1 provides a very specific and efficient therapeutic target for antisense oligonucleotide-mediated correction of SMN2 splicing in SMA.
Figures
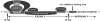



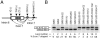
References
-
- Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251-256. - PubMed
-
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. - PubMed
-
- Buratti, E., and F. E. Baralle. 2005. Another step forward for SELEXive splicing. Trends Mol. Med. 11:5-9. - PubMed
-
- Cartegni, L., and A. R. Krainer. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30:377-384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
