Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast
- PMID: 16449648
- PMCID: PMC1367198
- DOI: 10.1128/MCB.26.4.1355-1372.2006
Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast
Abstract
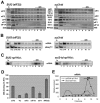
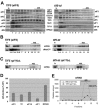
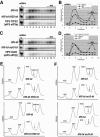
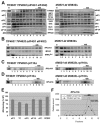
Recruitment of the eukaryotic translation initiation factor 2 (eIF2)-GTP-Met-tRNAiMet ternary complex to the 40S ribosome is stimulated by multiple initiation factors in vitro, including eIF3, eIF1, eIF5, and eIF1A. Recruitment of mRNA is thought to require the functions of eIF4F and eIF3, with the latter serving as an adaptor between the ribosome and the 4G subunit of eIF4F. To define the factor requirements for these reactions in vivo, we examined the effects of depleting eIF2, eIF3, eIF5, or eIF4G in Saccharomyces cerevisiae cells on binding of the ternary complex, other initiation factors, and RPL41A mRNA to native 43S and 48S preinitiation complexes. Depleting eIF2, eIF3, or eIF5 reduced 40S binding of all constituents of the multifactor complex (MFC), comprised of these three factors and eIF1, supporting a mechanism of coupled 40S binding by MFC components. 40S-bound mRNA strongly accumulated in eIF5-depleted cells, even though MFC binding to 40S subunits was reduced by eIF5 depletion. Hence, stimulation of the GTPase activity of the ternary complex, a prerequisite for 60S subunit joining in vitro, is likely the rate-limiting function of eIF5 in vivo. Depleting eIF2 or eIF3 impaired mRNA binding to free 40S subunits, but depleting eIF4G led unexpectedly to accumulation of mRNA on 40S subunits. Thus, it appears that eIF3 and eIF2 are more critically required than eIF4G for stable binding of at least some mRNAs to native preinitiation complexes and that eIF4G has a rate-limiting function at a step downstream of 48S complex assembly in vivo.
Figures

Similar articles
-
Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation.EMBO J. 2001 May 1;20(9):2326-37. doi: 10.1093/emboj/20.9.2326. EMBO J. 2001. PMID: 11331597 Free PMC article.
-
Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control.EMBO J. 2004 Mar 10;23(5):1166-77. doi: 10.1038/sj.emboj.7600116. Epub 2004 Feb 19. EMBO J. 2004. PMID: 14976554 Free PMC article.
-
A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo.Genes Dev. 2000 Oct 1;14(19):2534-46. doi: 10.1101/gad.831800. Genes Dev. 2000. PMID: 11018020 Free PMC article.
-
Molecular mechanisms of translation initiation in eukaryotes.Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7029-36. doi: 10.1073/pnas.111145798. Proc Natl Acad Sci U S A. 2001. PMID: 11416183 Free PMC article. Review.
-
The scanning mechanism of eukaryotic translation initiation.Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review.
Cited by
-
Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode.Mol Cell Biol. 2012 Oct;32(19):3978-89. doi: 10.1128/MCB.00376-12. Epub 2012 Jul 30. Mol Cell Biol. 2012. PMID: 22851688 Free PMC article.
-
Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit.J Biol Chem. 2006 Aug 11;281(32):22917-32. doi: 10.1074/jbc.M605418200. Epub 2006 Jun 9. J Biol Chem. 2006. PMID: 16766523 Free PMC article.
-
Control of Translation at the Initiation Phase During Glucose Starvation in Yeast.Int J Mol Sci. 2019 Aug 19;20(16):4043. doi: 10.3390/ijms20164043. Int J Mol Sci. 2019. PMID: 31430885 Free PMC article. Review.
-
Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining.Mol Cell Biol. 2007 Mar;27(6):2384-97. doi: 10.1128/MCB.02254-06. Epub 2007 Jan 22. Mol Cell Biol. 2007. PMID: 17242201 Free PMC article.
-
Transcriptional profile of breast muscle in heat stressed layers is similar to that of broiler chickens at control temperature.Genet Sel Evol. 2017 Sep 20;49(1):69. doi: 10.1186/s12711-017-0346-x. Genet Sel Evol. 2017. PMID: 28931372 Free PMC article.
References
-
- Asano, K., T. G. Kinzy, W. C. Merrick, and J. W. B. Hershey. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem. 272:1101-1109. - PubMed
-
- Asano, K., L. Phan, L. Valasek, L. W. Schoenfeld, A. Shalev, J. Clayton, K. Nielsen, T. F. Donahue, and A. G. Hinnebusch. 2001. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harbor Symp. Quant. Biol. 66:403-415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
