Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae
- PMID: 16449659
- PMCID: PMC1367203
- DOI: 10.1128/MCB.26.4.1496-1509.2006
Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae
Abstract
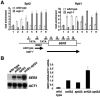
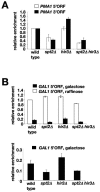
Spt2/Sin1 is a DNA binding protein with HMG-like domains that has been suggested to play a role in chromatin-mediated transcription in Saccharomyces cerevisiae. Previous studies have suggested models in which Spt2 plays an inhibitory role in the initiation of transcription of certain genes. In this work, we have taken several approaches to study Spt2 in greater detail. Our results have identified previously unknown genetic interactions between spt2Delta and mutations in genes encoding transcription elongation factors, including members of the PAF and HIR/HPC complexes. In addition, genome-wide and gene-specific chromatin immunoprecipitation analyses suggest that Spt2 is primarily associated with coding regions in a transcription-dependent fashion. Furthermore, our results show that Spt2, like other elongation factors, is required for the repression of transcription from a cryptic promoter within a coding region and that Spt2 is also required for repression of recombination within transcribed regions. Finally, we provide evidence that Spt2 plays a role in regulating the levels of histone H3 over transcribed regions. Taken together, our results suggest a direct link for Spt2 with transcription elongation, chromatin dynamics, and genome stability.
Figures






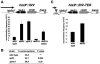
Similar articles
-
The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast.J Biol Chem. 2008 Oct 10;283(41):27350-27354. doi: 10.1074/jbc.C800147200. Epub 2008 Aug 15. J Biol Chem. 2008. PMID: 18708354
-
Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II.Mol Cell Biol. 2011 Mar;31(6):1288-300. doi: 10.1128/MCB.01083-10. Epub 2011 Jan 10. Mol Cell Biol. 2011. PMID: 21220514 Free PMC article.
-
Transcription elongation factors repress transcription initiation from cryptic sites.Science. 2003 Aug 22;301(5636):1096-9. doi: 10.1126/science.1087374. Science. 2003. PMID: 12934008
-
Insights into Spt6: a histone chaperone that functions in transcription, DNA replication, and genome stability.Trends Genet. 2023 Nov;39(11):858-872. doi: 10.1016/j.tig.2023.06.008. Epub 2023 Jul 20. Trends Genet. 2023. PMID: 37481442 Free PMC article. Review.
-
Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization.Mol Cells. 2014 Dec 31;37(12):851-6. doi: 10.14348/molcells.2014.0258. Epub 2014 Nov 5. Mol Cells. 2014. PMID: 25377252 Free PMC article. Review.
Cited by
-
A novel gene, Le-Dd10, is involved in fruiting body formation of Lentinula edodes.Arch Microbiol. 2022 Sep 5;204(10):602. doi: 10.1007/s00203-022-03206-z. Arch Microbiol. 2022. PMID: 36063239 Free PMC article.
-
Unique and Shared Roles for Histone H3K36 Methylation States in Transcription Regulation Functions.Cell Rep. 2020 Jun 9;31(10):107751. doi: 10.1016/j.celrep.2020.107751. Cell Rep. 2020. PMID: 32521276 Free PMC article.
-
Casein kinase 2 mediated phosphorylation of Spt6 modulates histone dynamics and regulates spurious transcription.Nucleic Acids Res. 2018 Sep 6;46(15):7612-7630. doi: 10.1093/nar/gky515. Nucleic Acids Res. 2018. PMID: 29905868 Free PMC article.
-
Regulation of chaperone binding and nucleosome dynamics by key residues within the globular domain of histone H3.Epigenetics Chromatin. 2016 Apr 30;9:17. doi: 10.1186/s13072-016-0066-4. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27134679 Free PMC article.
-
Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Aug;27(15):5575-86. doi: 10.1128/MCB.00459-07. Epub 2007 May 25. Mol Cell Biol. 2007. PMID: 17526727 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
-
- Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
