Comparison of microsatellites, single-nucleotide polymorphisms (SNPs) and composite markers derived from SNPs in linkage analysis
- PMID: 16451638
- PMCID: PMC1866757
- DOI: 10.1186/1471-2156-6-S1-S29
Comparison of microsatellites, single-nucleotide polymorphisms (SNPs) and composite markers derived from SNPs in linkage analysis
Abstract
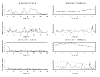
There is growing evidence that a map of dense single-nucleotide polymorphisms (SNPs) can outperform a map of sparse microsatellites for linkage analysis. There is also argument as to whether a clustered SNP map can outperform an evenly spaced SNP map. Using Genetic Analysis Workshop 14 simulated data, we compared for linkage analysis microsatellites, SNPs, and composite markers derived from SNPs. We encoded the composite markers in a two-step approach, in which the maximum identity length contrast method was employed to allow for recombination between loci. A SNP map 2.3 times as dense as a microsatellite map (approximately 2.9 cM compared to approximately 6.7 cM apart) provided slightly less information content (approximately 0.83 compared to approximately 0.89). Most inheritance information could be extracted when the SNPs were spaced < 1 cM apart. Comparing the linkage results on using SNPs or composite markers derived from them based on both 3 cM and 0.3 cM resolution maps, we showed that the inter-SNP distance should be kept small (< 1 cM), and that for multipoint linkage analysis the original markers and the derived composite markers had similar power; but for single point linkage analysis the resulting composite markers lead to more power. Considering all factors, such as information content, flexibility of analysis method, map errors, and genotyping errors, a map of clustered SNPs can be an efficient design for a genome-wide linkage scan.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

