Optimizing the evidence for linkage by permuting marker order
- PMID: 16451674
- PMCID: PMC1866721
- DOI: 10.1186/1471-2156-6-S1-S61
Optimizing the evidence for linkage by permuting marker order
Abstract
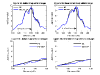
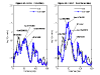
We developed a new marker-reordering algorithm to find the best order of fine-mapping markers for multipoint linkage analysis. The algorithm searches for the best order of fine-mapping markers such that the sum of the squared differences in identity-by-descent distribution between neighboring markers is minimized. To test this algorithm, we examined its effect on the evidence for linkage in the simulated and the Collaborative Studies on Genetics of Alcoholism (COGA) data. We found enhanced evidence for linkage with the reordered map at the true location in the simulated data (p-value decreased from 1.16 x 10(-9) to 9.70 x 10(-10)). Analysis of the White population from the COGA data with the reordered map for alcohol dependence led to a significant change of the linkage signal (p = 0.0365 decreased to p = 0.0039) on chromosome 1 between marker D1S1592 and D1S1598. Our results suggest that reordering fine-mapping markers in candidate regions when the genetic map is uncertain can be a critical step when considering a dense map.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

