Efficient gene targeting in Drosophila with zinc-finger nucleases
- PMID: 16452139
- PMCID: PMC1456366
- DOI: 10.1534/genetics.105.052829
Efficient gene targeting in Drosophila with zinc-finger nucleases
Abstract
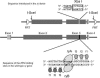
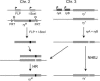
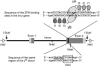
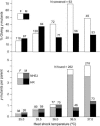
This report describes high-frequency germline gene targeting at two genomic loci in Drosophila melanogaster, y and ry. In the best case, nearly all induced parents produced mutant progeny; 25% of their offspring were new mutants and most of these were targeted gene replacements resulting from homologous recombination (HR) with a marked donor DNA. The procedure that generates these high frequencies relies on cleavage of the target by designed zinc-finger nucleases (ZFNs) and production of a linear donor in situ. Increased induction of ZFN expression led to higher frequencies of gene targeting, demonstrating the beneficial effect of activating the target. In the absence of a homologous donor DNA, ZFN cleavage led to the recovery of new mutants at three loci-y, ry and bw-through nonhomologous end joining (NHEJ) after cleavage. Because zinc fingers can be directed to a broad range of DNA sequences and targeting is very efficient, this approach promises to allow genetic manipulation of many different genes, even in cases where the mutant phenotype cannot be predicted.
Figures

References
-
- Alwin, S., M. B. Gere, E. Gulh, K. Effertz, C. F. Barbas, III et al., 2005. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 12: 610–617. - PubMed
-
- Ashburner, M., K. G. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Beall, E. L., and D. C. Rio, 1996. Drosophila IRBP/Ku70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 10: 921–933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

