Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli
- PMID: 16452142
- PMCID: PMC1456380
- DOI: 10.1534/genetics.105.052076
Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli
Abstract
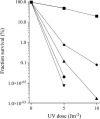
Heterotrimeric RecBCD enzyme unwinds and resects a DNA duplex containing blunt double-stranded ends and directs loading of the strand-exchange protein RecA onto the unwound 3'-ending strand, thereby initiating the majority of recombination in wild-type Escherichia coli. When the enzyme lacks its RecD subunit, the resulting RecBC enzyme, active in recD mutants, is recombination proficient although it has only helicase and RecA loading activity and is not a nuclease. However, E. coli encodes for several other exonucleases that digest double-stranded and single-stranded DNA and thus might act in consort with the RecBC enzyme to efficiently promote recombination reactions. To test this hypothesis, I inactivated multiple exonucleases (i.e., exonuclease I, exonuclease X, exonuclease VII, RecJ, and SbcCD) in recD derivatives of the wild-type and nuclease-deficient recB1067 strain and assessed the ability of the resultant mutants to maintain cell viability and to promote DNA repair and homologous recombination. A complex pattern of overlapping and sometimes competing activities of multiple exonucleases in recD mutants was thus revealed. These exonucleases were shown to be essential for cell viability, DNA repair (of UV- and gamma-induced lesions), and homologous recombination (during Hfr conjugation and P1 transduction), which are dependent on the RecBC enzyme. A model for donor DNA processing in recD transconjugants and transductants was proposed.
Figures
Similar articles
-
Effects of recJ, recQ, and recFOR mutations on recombination in nuclease-deficient recB recD double mutants of Escherichia coli.J Bacteriol. 2005 Feb;187(4):1350-6. doi: 10.1128/JB.187.4.1350-1356.2005. J Bacteriol. 2005. PMID: 15687199 Free PMC article.
-
The hybrid recombinational repair pathway operates in a χ activity deficient recC1004 mutant of Escherichia coli.Biochimie. 2012 Sep;94(9):1918-25. doi: 10.1016/j.biochi.2012.05.008. Epub 2012 May 19. Biochimie. 2012. PMID: 22617484
-
Exonuclease requirements for recombination of lambda-phage in recD mutants of Escherichia coli.Genetics. 2006 Aug;173(4):2399-402. doi: 10.1534/genetics.106.060426. Epub 2006 May 15. Genetics. 2006. PMID: 16702415 Free PMC article. Review.
-
RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli.Res Microbiol. 2005 Apr;156(3):304-11. doi: 10.1016/j.resmic.2004.10.005. Epub 2004 Dec 1. Res Microbiol. 2005. PMID: 15808933
-
Chi and the RecBC D enzyme of Escherichia coli.Annu Rev Genet. 1994;28:49-70. doi: 10.1146/annurev.ge.28.120194.000405. Annu Rev Genet. 1994. PMID: 7893137 Review.
Cited by
-
Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease.Nucleic Acids Res. 2013 Aug;41(15):7556-65. doi: 10.1093/nar/gkt495. Epub 2013 Jun 14. Nucleic Acids Res. 2013. PMID: 23771145 Free PMC article.
-
Still finding ways to augment the existing management of acute and chronic kidney diseases with targeted gene and cell therapies: Opportunities and hurdles.Front Med (Lausanne). 2023 Mar 7;10:1143028. doi: 10.3389/fmed.2023.1143028. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36960337 Free PMC article. Review.
-
Homologous recombination mediated by the mycobacterial AdnAB helicase without end resection by the AdnAB nucleases.Nucleic Acids Res. 2017 Jan 25;45(2):762-774. doi: 10.1093/nar/gkw1130. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899634 Free PMC article.
-
Single-strand DNA-binding protein suppresses illegitimate recombination in Escherichia coli, acting in synergy with RecQ helicase.Sci Rep. 2024 Sep 3;14(1):20476. doi: 10.1038/s41598-024-70817-5. Sci Rep. 2024. PMID: 39227621 Free PMC article.
-
Roles of ExoI and SbcCD nucleases in "reckless" DNA degradation in recA mutants of Escherichia coli.J Bacteriol. 2009 Mar;191(5):1677-87. doi: 10.1128/JB.01877-07. Epub 2008 Dec 12. J Bacteriol. 2009. PMID: 19074388 Free PMC article.
References
-
- Anderson, D. G., and S. C. Kowalczykowski, 1997. a The recombination hot spot, Chi, is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11: 571–581. - PubMed
-
- Anderson, D. G., and S. C. Kowalczykowski, 1997. b The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90: 77–86. - PubMed
-
- Anderson, D. G., J. J. Churchill and S. C. Kowalczykowski, 1997. Chi-activated RecBCD enzyme possesses 5′-3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply the ejection of the RecD subunit. Genes Cells 2: 117–128. - PubMed
-
- Anderson, D. G., J. J. Churchill and S. C. Kowalczykowski, 1999. A single mutation, RecBD1080A, eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J. Biol. Chem. 274: 27139–27144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

