The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32
- PMID: 16452402
- PMCID: PMC1367211
- DOI: 10.1128/JB.188.4.1218-1226.2006
The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32
Abstract
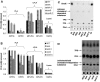
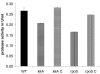
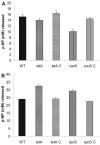
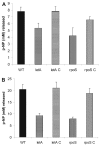
Legionella pneumophila possesses a variety of secreted and cell-associated hydrolytic activities that could be involved in pathogenesis. The activities include phospholipase A, lysophospholipase A, glycerophospholipid:cholesterol acyltransferase, lipase, protease, phosphatase, RNase, and p-nitrophenylphosphorylcholine (p-NPPC) hydrolase. Up to now, there have been no data available on the regulation of the enzymes in L. pneumophila and no data at all concerning the regulation of bacterial phospholipases A. Therefore, we used L. pneumophila mutants in the genes coding for the global regulatory proteins RpoS and LetA to investigate the dependency of hydrolytic activities on a global regulatory network proposed to control important virulence traits in L. pneumophila. Our results show that both L. pneumophila rpoS and letA mutants exhibit on the one hand a dramatic reduction of secreted phospholipase A and glycerophospholipid:cholesterol acyltransferase activities, while on the other hand secreted lysophospholipase A and lipase activities were significantly increased during late logarithmic growth phase. The cell-associated phospholipase A, lysophospholipase A, and p-NPPC hydrolase activities, as well as the secreted protease, phosphatase, and p-NPPC hydrolase activities were significantly decreased in both of the mutant strains. Only cell-associated phosphatase activity was slightly increased. In contrast, RNase activity was not affected. The expression of plaC, coding for a secreted acyltransferase, phospholipase A, and lysophospholipase A, was found to be regulated by LetA and RpoS. In conclusion, our results show that RpoS and LetA affect phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of L. pneumophila in a similar way, thereby corroborating the existence of the LetA/RpoS regulation cascade.
Figures

References
-
- Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

