Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress
- PMID: 16452403
- PMCID: PMC1367249
- DOI: 10.1128/JB.188.4.1227-1235.2006
Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress
Abstract
Proline is converted to glutamate in two successive steps by the proline utilization A (PutA) flavoenzyme in gram-negative bacteria. PutA contains a proline dehydrogenase domain that catalyzes the flavin adenine dinucleotide (FAD)-dependent oxidation of proline to delta1-pyrroline-5-carboxylate (P5C) and a P5C dehydrogenase domain that catalyzes the NAD+-dependent oxidation of P5C to glutamate. Here, we characterize PutA from Helicobacter hepaticus (PutA(Hh)) and Helicobacter pylori (PutA(Hp)) to provide new insights into proline metabolism in these gastrointestinal pathogens. Both PutA(Hh) and PutA(Hp) lack DNA binding activity, in contrast to PutA from Escherichia coli (PutA(Ec)), which both regulates and catalyzes proline utilization. PutA(Hh) and PutA(Hp) display catalytic activities similar to that of PutA(Ec) but have higher oxygen reactivity. PutA(Hh) and PutA(Hp) exhibit 100-fold-higher turnover numbers (approximately 30 min(-1)) than PutA(Ec) (<0. 3 min(-1)) using oxygen as an electron acceptor during catalytic turnover with proline. Consistent with increased oxygen reactivity, PutA(Hh) forms a reversible FAD-sulfite adduct. The significance of increased oxygen reactivity in PutA(Hh) and PutA(Hp) was probed by oxidative stress studies in E. coli. Expression of PutA(Ec) and PutA from Bradyrhizobium japonicum, which exhibit low oxygen reactivity, does not diminish stress survival rates of E. coli cell cultures. In contrast, PutA(Hp) and PutA(Hh) expression dramatically reduces E. coli cell survival and is correlated with relatively lower proline levels and increased hydrogen peroxide formation. The discovery of reduced oxygen species formation by PutA suggests that proline catabolism may influence redox homeostasis in the ecological niches of these Helicobacter species.
Figures
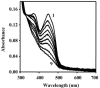
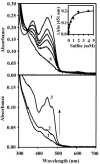

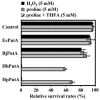
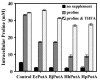
Similar articles
-
Characterization of a bifunctional PutA homologue from Bradyrhizobium japonicum and identification of an active site residue that modulates proline reduction of the flavin adenine dinucleotide cofactor.Biochemistry. 2005 Jun 28;44(25):9130-9. doi: 10.1021/bi050629k. Biochemistry. 2005. PMID: 15966737 Free PMC article.
-
The PutA protein of Salmonella typhimurium catalyzes the two steps of proline degradation via a leaky channel.Arch Biochem Biophys. 1998 Jun 15;354(2):281-7. doi: 10.1006/abbi.1998.0697. Arch Biochem Biophys. 1998. PMID: 9637737
-
Regulation of PutA-membrane associations by flavin adenine dinucleotide reduction.Biochemistry. 2004 Oct 19;43(41):13165-74. doi: 10.1021/bi048596g. Biochemistry. 2004. PMID: 15476410 Free PMC article.
-
Structure, function, and mechanism of proline utilization A (PutA).Arch Biochem Biophys. 2017 Oct 15;632:142-157. doi: 10.1016/j.abb.2017.07.005. Epub 2017 Jul 14. Arch Biochem Biophys. 2017. PMID: 28712849 Free PMC article. Review.
-
Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli.Amino Acids. 2008 Nov;35(4):711-8. doi: 10.1007/s00726-008-0053-6. Epub 2008 Mar 7. Amino Acids. 2008. PMID: 18324349 Free PMC article. Review.
Cited by
-
Elucidation of the Metabolic Network of Helicobacter pylori J99 and Malaysian Clinical Strains by Phenotype Microarray.Helicobacter. 2017 Feb;22(1):e12321. doi: 10.1111/hel.12321. Epub 2016 Jun 3. Helicobacter. 2017. PMID: 27258354 Free PMC article.
-
Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway.Genetics. 2013 Jun;194(2):421-33. doi: 10.1534/genetics.113.150326. Epub 2013 Apr 5. Genetics. 2013. PMID: 23564202 Free PMC article.
-
The ferric uptake regulator of Helicobacter pylori: a critical player in the battle for iron and colonization of the stomach.Future Microbiol. 2013 Jun;8(6):725-38. doi: 10.2217/fmb.13.43. Future Microbiol. 2013. PMID: 23701330 Free PMC article. Review.
-
RNA-Seq Profiling of Intestinal Expression of Xenobiotic Processing Genes in Germ-Free Mice.Drug Metab Dispos. 2017 Dec;45(12):1225-1238. doi: 10.1124/dmd.117.077313. Epub 2017 Sep 22. Drug Metab Dispos. 2017. PMID: 28939687 Free PMC article.
-
Proline dehydrogenase is a positive regulator of cell death in different kingdoms.Plant Signal Behav. 2011 Aug;6(8):1195-7. doi: 10.4161/psb.6.8.15791. Epub 2011 Aug 1. Plant Signal Behav. 2011. PMID: 21757996 Free PMC article.
References
-
- Abrahamson, J. L. A., L. G. Baker, J. T. Stephenson, and J. M. Wood. 1983. Proline dehydrogenase from Escherichia coli K12, properties of the membrane-associated enzyme. Eur. J. Biochem. 134:77-82. - PubMed
-
- Alamuri, P., and R. J. Maier. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:1397-1406. - PubMed
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Bates, L. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39:205-207.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

