CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6)
- PMID: 16452408
- PMCID: PMC1367233
- DOI: 10.1128/JB.188.4.1266-1278.2006
CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6)
Abstract
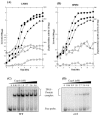
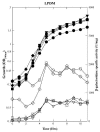
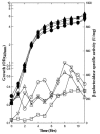
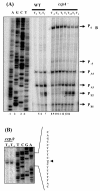
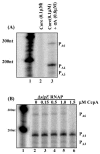
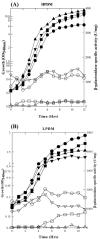
The Bacillus subtilis PhoPR two-component system is directly responsible for activation or repression of Pho regulon genes in response to phosphate deprivation. The response regulator, PhoP, and the histidine kinase, PhoR, are encoded in a single operon with a complex promoter region that contains five known transcription start sites, which respond to at least two regulatory proteins. We report here the identification of another direct regulator of phoPR transcription, carbon catabolite protein A, CcpA. This regulator functions in the presence of glucose or other readily metabolized carbon sources. The maximum derepression of phoPR expression in a ccpA mutant compared to a wild-type stain was observed under excess phosphate conditions with glucose either throughout growth in a high-phosphate defined medium or in a low-phosphate defined medium during exponential growth, a growth condition when phoPR transcription is low in a wild-type strain due to the absence of autoinduction. Either HPr or Crh were sufficient to cause CcpA dependent repression of the phoPR promoter in vivo. A ptsH1 (Hpr) crh double mutant completely relieves phoPR repression during phosphate starvation but not during phosphate replete growth. In vivo and in vitro studies showed that CcpA repressed phoPR transcription by binding directly to the cre consensus sequence present in the promoter. Primer extension and in vitro transcription studies revealed that the CcpA regulation of phoPR transcription was due to repression of P(A6), a previously unidentified promoter positioned immediately upstream of the cre box. Esigma(A) was sufficient for transcription of P(A6), which was repressed by CcpA in vitro. These studies showed direct repression by CcpA of a newly discovered Esigma(A)-responsive phoPR promoter that required either Hpr or Crh in vivo for direct binding to the putative consensus cre sequence located between P(A6) and the five downstream promoters characterized previously.
Figures



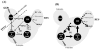
Similar articles
-
Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC.J Bacteriol. 2010 Jun;192(12):3103-13. doi: 10.1128/JB.00089-10. Epub 2010 Apr 9. J Bacteriol. 2010. PMID: 20382764 Free PMC article.
-
Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP.J Bacteriol. 2004 Jul;186(13):4262-75. doi: 10.1128/JB.186.13.4262-4275.2004. J Bacteriol. 2004. PMID: 15205429 Free PMC article.
-
Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response.J Bacteriol. 2005 Aug;187(15):5166-78. doi: 10.1128/JB.187.15.5166-5178.2005. J Bacteriol. 2005. PMID: 16030210 Free PMC article.
-
The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis.FEMS Microbiol Lett. 1996 Jan 1;135(1):9-15. doi: 10.1111/j.1574-6968.1996.tb07959.x. FEMS Microbiol Lett. 1996. PMID: 8598282 Review.
-
Carbon catabolite control of the metabolic network in Bacillus subtilis.Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review.
Cited by
-
Genomic Analysis of Microbulbifer sp. Strain A4B-17 and the Characterization of Its Metabolic Pathways for 4-Hydroxybenzoic Acid Synthesis.Front Microbiol. 2018 Dec 18;9:3115. doi: 10.3389/fmicb.2018.03115. eCollection 2018. Front Microbiol. 2018. PMID: 30619190 Free PMC article.
-
Regulators of the Bacillus subtilis cydABCD operon: identification of a negative regulator, CcpA, and a positive regulator, ResD.J Bacteriol. 2007 May;189(9):3348-58. doi: 10.1128/JB.00050-07. Epub 2007 Feb 23. J Bacteriol. 2007. PMID: 17322317 Free PMC article.
-
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.BMC Genomics. 2012 Aug 17;13:401. doi: 10.1186/1471-2164-13-401. BMC Genomics. 2012. PMID: 22900538 Free PMC article.
-
Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools.J Bacteriol. 2010 Dec;192(24):6357-68. doi: 10.1128/JB.00937-10. Epub 2010 Oct 8. J Bacteriol. 2010. PMID: 20935095 Free PMC article.
-
Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.Nucleic Acids Res. 2011 Apr;39(7):2931-42. doi: 10.1093/nar/gkq1177. Epub 2010 Nov 23. Nucleic Acids Res. 2011. PMID: 21106498 Free PMC article.
References
-
- Birkey, S. M. 1998. Regulation of the phoPR operon of Bacillus subtilis during phosphate starvation and sporulation. Ph.D. thesis. University of Illinois at Chicago, Chicago, Ill.
-
- Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

