The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli
- PMID: 16452414
- PMCID: PMC1367212
- DOI: 10.1128/JB.188.4.1316-1331.2006
The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli
Abstract
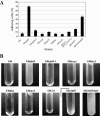
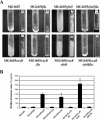
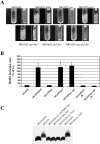
We investigated the influence of regulatory and pathogenicity island-associated factors (Hha, RpoS, LuxS, EvgA, RfaH, and tRNA5Leu) on biofilm formation by uropathogenic Escherichia coli (UPEC) strain 536. Only inactivation of rfaH, which encodes a transcriptional antiterminator, resulted in increased initial adhesion and biofilm formation by E. coli 536. rfaH inactivation in nonpathogenic E. coli K-12 isolate MG1655 resulted in the same phenotype. Transcriptome analysis of wild-type strain 536 and an rfaH mutant of this strain revealed that deletion of rfaH correlated with increased expression of flu orthologs. flu encodes antigen 43 (Ag43), which mediates autoaggregation and biofilm formation. We confirmed that deletion of rfaH leads to increased levels of flu and flu-like transcripts in E. coli K-12 and UPEC. Supporting the hypothesis that RfaH represses biofilm formation through reduction of the Ag43 level, the increased-biofilm phenotype of E. coli MG1655rfaH was reversed upon inactivation of flu. Deletion of the two flu orthologs, however, did not modify the behavior of mutant 536rfaH. Our results demonstrate that the strong initial adhesion and biofilm formation capacities of strain MG1655rfaH are mediated by both increased steady-state production of Ag43 and likely increased Ag43 presentation due to null rfaH-dependent lipopolysaccharide depletion. Although the roles of rfaH in the biofilm phenotype are different in UPEC strain 536 and K-12 strain MG1655, this study shows that RfaH, in addition to affecting the expression of bacterial virulence factors, also negatively controls expression and surface presentation of Ag43 and possibly another Ag43-independent factor(s) that mediates cell-cell interactions and biofilm formation.
Figures


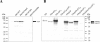

References
-
- Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. - PubMed
-
- Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. - PubMed
-
- Bailey, M. J., C. Hughes, and V. Koronakis. 2000. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol. Gen. Genet. 262:1052-1059. - PubMed
-
- Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

