The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1
- PMID: 16452425
- PMCID: PMC1367250
- DOI: 10.1128/JB.188.4.1419-1436.2006
The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1
Abstract
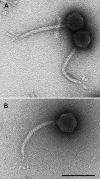
A new Escherichia coli phage, named Rtp, was isolated and shown to be closely related to phage T1. Electron microscopy revealed that phage Rtp has a morphologically unique tail tip consisting of four leaf-like structures arranged in a rosette, whereas phage T1 has thinner, flexible leaves that thicken toward the ends. In contrast to T1, Rtp did not require FhuA and TonB for infection. The 46.2-kb genome of phage Rtp encodes 75 open reading frames, 47 of which are homologous to phage T1 genes. Like phage T1, phage Rtp encodes a large number of small genes at the genome termini that exhibit no sequence similarity to known genes. Six predicted genes larger than 300 nucleotides in the highly homologous region of Rtp are not found in T1. Two predicted HNH endonucleases are encoded at positions different from those in phage T1. The sequence similarity of rtp37, -38, -39, -41, -42, and -43 to equally arranged genes of lambdoid phages suggests a common tail assembly initiation complex. Protein Rtp43 is homologous to the lambda J protein, which determines lambda host specificity. Since the two proteins differ most in the C-proximal area, where the binding site to the LamB receptor resides in the J protein, we propose that Rtp43 contributes to Rtp host specificity. Lipoproteins similar to the predicted lipoprotein Rtp45 are found in a number of phages (encoded by cor genes) in which they prevent superinfection by inactivating the receptors. We propose that, similar to the proposed function of the phage T5 lipoprotein, Rtp45 prevents inactivation of Rtp by adsorption to its receptor during cells lysis. Rtp52 is a putative transcriptional regulator, for which 10 conserved inverted repeats were identified upstream of genes in the Rtp genome. In contrast, the much larger E. coli genome has only one such repeat sequence.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

