The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans
- PMID: 16452431
- PMCID: PMC1367237
- DOI: 10.1128/JB.188.4.1473-1488.2006
The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans
Abstract
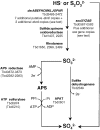
The complete genome sequence of Thiobacillus denitrificans ATCC 25259 is the first to become available for an obligately chemolithoautotrophic, sulfur-compound-oxidizing, beta-proteobacterium. Analysis of the 2,909,809-bp genome will facilitate our molecular and biochemical understanding of the unusual metabolic repertoire of this bacterium, including its ability to couple denitrification to sulfur-compound oxidation, to catalyze anaerobic, nitrate-dependent oxidation of Fe(II) and U(IV), and to oxidize mineral electron donors. Notable genomic features include (i) genes encoding c-type cytochromes totaling 1 to 2 percent of the genome, which is a proportion greater than for almost all bacterial and archaeal species sequenced to date, (ii) genes encoding two [NiFe]hydrogenases, which is particularly significant because no information on hydrogenases has previously been reported for T. denitrificans and hydrogen oxidation appears to be critical for anaerobic U(IV) oxidation by this species, (iii) a diverse complement of more than 50 genes associated with sulfur-compound oxidation (including sox genes, dsr genes, and genes associated with the AMP-dependent oxidation of sulfite to sulfate), some of which occur in multiple (up to eight) copies, (iv) a relatively large number of genes associated with inorganic ion transport and heavy metal resistance, and (v) a paucity of genes encoding organic-compound transporters, commensurate with obligate chemolithoautotrophy. Ultimately, the genome sequence of T. denitrificans will enable elucidation of the mechanisms of aerobic and anaerobic sulfur-compound oxidation by beta-proteobacteria and will help reveal the molecular basis of this organism's role in major biogeochemical cycles (i.e., those involving sulfur, nitrogen, and carbon) and groundwater restoration.
Figures

References
-
- Aminuddin, M., and D. J. D. Nicholas. 1974. An AMP-independent sulphite oxidase from Thiobacillus denitrificans: purification and properties. J. Gen. Microbiol. 82:103-113.
-
- Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. - PubMed
-
- Beijerinck, M. W. 1904. Phénomènes de réduction produits par les microbes (Conférence avec demonstrations faite—Delft, le 16 avril 1903). Arch. Neerl. Sci. Ser. 2 9:131-157.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

