In the Bacillus stearothermophilus DnaB-DnaG complex, the activities of the two proteins are modulated by distinct but overlapping networks of residues
- PMID: 16452437
- PMCID: PMC1367256
- DOI: 10.1128/JB.188.4.1534-1539.2006
In the Bacillus stearothermophilus DnaB-DnaG complex, the activities of the two proteins are modulated by distinct but overlapping networks of residues
Abstract
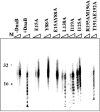
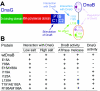
We demonstrate the primase activity of Bacillus stearothermophilus DnaG and show that it initiates at 3'-ATC-5' and 3'-ATT-5' sites synthesizing primers that are 22 or 23 nucleotides long. In the presence of the helicase DnaB the size distribution of primers is different, and a range of additional smaller primers are also synthesized. Nine residues from the N- and C-terminal domains of DnaB, as well as its linker region, have been reported previously to affect this interaction. In Bacillus stearothermophilus only three residues from the linker region (I119 and I125) and the N-terminal domain (Y88) of DnaB have been shown previously to have direct structural importance, and I119 and I125 mediate DnaG-induced effects on DnaB activity. The functions of the other residues (L138, T191, E192, R195, and M196) are still a mystery. Here we show that the E15A, Y88A, and E15A Y88A mutants bind DnaG but are not able to modulate primer size, whereas the R195A M196A mutant inhibited the primase activity. Therefore, four of these residues, E15 and Y88 (N-terminal domain) and R195 and M196 (C-terminal domain), mediate DnaB-induced effects on DnaG activity. Overall, the data suggest that the effects of DnaB on DnaG activity and vice versa are mediated by distinct but overlapping networks of residues.
Figures


References
-
- Bhattacharyya, S., and M. A. Griep. 2000. DnaB helicase affects the initiation specificity of Escherichia coli primase on single-stranded DNA templates. Biochemistry 39:745-752. - PubMed
-
- Chang, P., and K. J. Marians. 2000. Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J. Biol. Chem. 275:26187-26195. - PubMed
-
- Corn, J. E., P. J. Pease, G. L. Hura, and J. M. Berger. 2005. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20:391-401. - PubMed
-
- Fang, L., M. J. Davey, and M. O'Donnell. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541-553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

