Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243
- PMID: 16452439
- PMCID: PMC1367220
- DOI: 10.1128/JB.188.4.1551-1566.2006
Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243
Abstract
Burkholderia pseudomallei is a gram-negative facultative intracellular pathogen that causes melioidosis, an invasive disease of humans and animals. To address the response of this bacterium to iron-limiting conditions, we first performed a global transcriptional analysis of RNA extracted from bacteria grown under iron-limiting and iron-rich conditions by microarrays. We focused our study on those open reading frames (ORFs) induced under iron limitation, which encoded predicted proteins that could be involved in the biosynthesis and uptake of the siderophore malleobactin. We purified this siderophore and determined that it consisted of at least three compounds with different molecular weights. We demonstrated that ORFs BPSL1776 and BPSL1774, designated mbaA and mbaF, respectively, are involved in the biosynthesis of malleobactin, while BPSL1775, named fmtA, is involved in its transport. These genes are in an operon with two other ORFs (mbaJ and mbaI) whose transcription is under the control of MbaS, a protein that belongs to the extracytoplasmic function sigma factors. Interestingly, the transcription of the mbaA, fmtA, and mbaS genes is not controlled by the availability of the siderophore malleobactin.
Figures


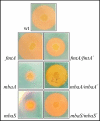

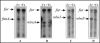


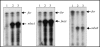
References
-
- Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

