The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins
- PMID: 16452444
- PMCID: PMC1367222
- DOI: 10.1128/JB.188.4.1607-1614.2006
The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins
Abstract
The elaC gene product from Escherichia coli, ZiPD, is a 3' tRNA-processing endonuclease belonging to the tRNase Z family of enzymes that have been identified in a wide variety of organisms. In contrast to the elaC homologue from Bacillus subtilis, E. coli elaC is not essential for viability, and although both enzymes process only precursor tRNA (pre-tRNA) lacking a CCA triplet at the 3' end in vitro, the physiological role of ZiPD remains enigmatic because all pre-tRNA species in E. coli are transcribed with the CCA triplet. We present the first crystal structure of ZiPD determined by multiple anomalous diffraction at a resolution of 2.9 A. This structure shares many features with the tRNase Z enzymes from B. subtilis and Thermotoga maritima, but there are distinct differences in metal binding and overall domain organization. Unlike the previously described homologous structures, ZiPD dimers display crystallographic symmetry and fully loaded metal sites. The ZiPD exosite is similar to that of the B. subtilis enzyme structurally, but its position with respect to the protein core differs substantially, illustrating its ability to act as a clamp in binding tRNA. Furthermore, the ZiPD crystal structure presented here provides insight into the enzyme's cooperativity and assists the ongoing attempt to elucidate the physiological function of this protein.
Figures
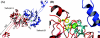




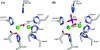
Similar articles
-
Exosite modules guide substrate recognition in the ZiPD/ElaC protein family.J Biol Chem. 2005 May 6;280(18):17857-62. doi: 10.1074/jbc.M500591200. Epub 2005 Feb 7. J Biol Chem. 2005. PMID: 15699034
-
Characterization of an Escherichia coli elaC deletion mutant.Biochem Biophys Res Commun. 2004 Aug 6;320(4):1365-73. doi: 10.1016/j.bbrc.2004.05.227. Biochem Biophys Res Commun. 2004. PMID: 15303284
-
The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Aug 1;63(Pt 8):637-41. doi: 10.1107/S1744309107033623. Epub 2007 Jul 21. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007. PMID: 17671357 Free PMC article.
-
Zinc- and iron-dependent cytosolic metallo-beta-lactamase domain proteins exhibit similar zinc-binding affinities, independent of an atypical glutamate at the metal-binding site.Biochem J. 2005 Jan 1;385(Pt 1):145-53. doi: 10.1042/BJ20040773. Biochem J. 2005. PMID: 15324305 Free PMC article.
-
The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties.Biol Chem. 2005 Dec;386(12):1253-64. doi: 10.1515/BC.2005.142. Biol Chem. 2005. PMID: 16336119 Review.
Cited by
-
tRNase Z catalysis and conserved residues on the carboxy side of the His cluster.Biochemistry. 2007 Aug 21;46(33):9380-7. doi: 10.1021/bi700578v. Epub 2007 Jul 27. Biochemistry. 2007. PMID: 17655328 Free PMC article.
-
Identification by Mn2+ rescue of two residues essential for the proton transfer of tRNase Z catalysis.Nucleic Acids Res. 2006 Aug 11;34(13):3811-8. doi: 10.1093/nar/gkl517. Print 2006. Nucleic Acids Res. 2006. PMID: 16916792 Free PMC article.
-
tRNA biology charges to the front.Genes Dev. 2010 Sep 1;24(17):1832-60. doi: 10.1101/gad.1956510. Genes Dev. 2010. PMID: 20810645 Free PMC article. Review.
-
Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation.J Biol Chem. 2009 Jun 19;284(25):17216-17226. doi: 10.1074/jbc.M808392200. Epub 2009 Apr 14. J Biol Chem. 2009. PMID: 19366688 Free PMC article.
-
Characterization of the Methanomicrobial Archaeal RNase Zs for Processing the CCA-Containing tRNA Precursors.Front Microbiol. 2020 Aug 25;11:1851. doi: 10.3389/fmicb.2020.01851. eCollection 2020. Front Microbiol. 2020. PMID: 32982996 Free PMC article.
References
-
- Aravind, L. 1999. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 1:69-91. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Brunger, A. T. 1993. Assessment of phase accuracy by cross validation—the free R-value—methods and applications. Acta Crystallogr. Sect. D Biol. Crystallogr. 49:24-36. - PubMed
-
- Brunger, A. T. 1992. Free R-value—a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

