Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1
- PMID: 16452505
- PMCID: PMC1361704
- DOI: 10.1101/gad.1380506
Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1
Abstract
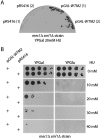
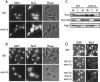
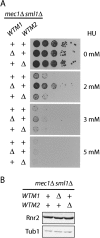
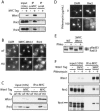
The control of deoxyribonucleotide levels is essential for DNA synthesis and repair. This control is exerted through regulation of ribonucleotide reductase (RNR). One mode of RNR regulation is differential localization of its subunits. In Saccharomyces cerevisiae, the catalytic subunit hererodimer, Rnr2/Rnr4, is localized to the nucleus while its regulatory subunit, Rnr1, is cytoplasmic. During S phase and in response to DNA damage, Rnr2-Rnr4 enters the cytoplasm, where it presumably combines with Rnr1 to form an active complex. The mechanism of its nuclear localization is not understood. Here, we report the isolation of the WTM (WD40-containing transcriptional modulator) proteins as regulators of Rnr2/Rnr4 localization. Overproduction of Wtm2 increased Rnr2/Rnr4. Deletion of WTM1, a homolog of WTM2, leads to the cytoplasmic localization of Rnr2/Rnr4, and increased hydroxyurea (HU)-resistance in mec1 mutants. Wtm1 binds Rnr2/4 complexes and release them to the cytoplasm in response to DNA damage. Forced localization of Wtm1 to the nucleolus causes Rnr2/Rnr4 complexes to relocalize to the nucleolus. Thus, Wtm1 acts as a nuclear anchor to maintain nuclear localization of Rnr2/4 complexes outside of S phase. In the presence of DNA damage this association is disrupted and Rnr2/Rnr4 become cytoplasmic, where they join with Rnr1 to form an intact complex.
Figures

Similar articles
-
Yeast Dun1 Kinase Regulates Ribonucleotide Reductase Small Subunit Localization in Response to Iron Deficiency.J Biol Chem. 2016 Apr 29;291(18):9807-17. doi: 10.1074/jbc.M116.720862. Epub 2016 Mar 12. J Biol Chem. 2016. PMID: 26970775 Free PMC article.
-
Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase.Mol Cell. 2008 Oct 10;32(1):70-80. doi: 10.1016/j.molcel.2008.08.018. Mol Cell. 2008. PMID: 18851834 Free PMC article.
-
Clb6-Cdc28 Promotes Ribonucleotide Reductase Subcellular Redistribution during S Phase.Mol Cell Biol. 2018 Feb 27;38(6):e00497-17. doi: 10.1128/MCB.00497-17. Print 2018 Mar 15. Mol Cell Biol. 2018. PMID: 29263158 Free PMC article.
-
DNA damage and cell cycle regulation of ribonucleotide reductase.Bioessays. 1993 May;15(5):333-9. doi: 10.1002/bies.950150507. Bioessays. 1993. PMID: 8343143 Review.
-
Rnr1's role in telomere elongation cannot be replaced by Rnr3: a role beyond dNTPs?Curr Genet. 2018 Jun;64(3):547-550. doi: 10.1007/s00294-017-0779-3. Epub 2017 Nov 8. Curr Genet. 2018. PMID: 29119271 Review.
Cited by
-
Yeast Dun1 Kinase Regulates Ribonucleotide Reductase Small Subunit Localization in Response to Iron Deficiency.J Biol Chem. 2016 Apr 29;291(18):9807-17. doi: 10.1074/jbc.M116.720862. Epub 2016 Mar 12. J Biol Chem. 2016. PMID: 26970775 Free PMC article.
-
Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases.Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10364-9. doi: 10.1073/pnas.0701622104. Epub 2007 Jun 11. Proc Natl Acad Sci U S A. 2007. PMID: 17563356 Free PMC article.
-
Regulation of DNA duplication by the mTOR signaling pathway.Cell Cycle. 2021 Apr;20(8):742-751. doi: 10.1080/15384101.2021.1897271. Epub 2021 Mar 10. Cell Cycle. 2021. PMID: 33691584 Free PMC article. Review.
-
Essential functions of iron-requiring proteins in DNA replication, repair and cell cycle control.Protein Cell. 2014 Oct;5(10):750-60. doi: 10.1007/s13238-014-0083-7. Epub 2014 Jul 8. Protein Cell. 2014. PMID: 25000876 Free PMC article. Review.
-
The available SRL3 deletion strain of Saccharomyces cerevisiae contains a truncation of DNA damage tolerance protein Mms2: Implications for Srl3 and Mms2 functions.Internet J Microbiol. 2009;8(1):8. doi: 10.5580/42c. Internet J Microbiol. 2009. PMID: 24795789 Free PMC article.
References
-
- Allen J.B., Zhou, Z., Siede, W., Friedberg, E.C., and Elledge, S.J. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes & Dev. 8: 2401-2415. - PubMed
-
- Bjorklund S., Skog, S., Tribukait, B., and Thelander, L. 1990. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 29: 5452-5458. - PubMed
-
- Bonifacino J.S., Dell'Angelical, E.C., and Springer, T.A. 1999. Immunoprecipitation. In Current protocols in protein science (eds. J.E. Coligan et al.), Unit 9.8. Wiley Interscience, Hoboken, NJ. - PubMed
-
- Brissenden J.E., Caras, I., Thelander, L., and Francke, U. 1988. The structural gene for the M1 subunit of ribonucleotide reductase maps to chromosome 11, band p15, in human and to chromosome 7 in mouse. Exp. Cell Res. 174: 302-308. - PubMed
-
- Chabes A., Georgieva, B., Domkin, V., Zhao, X., Rothstein, R., and Thelander, L. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391-401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
