Mimicking the action of GroEL in molecular dynamics simulations: application to the refinement of protein structures
- PMID: 16452612
- PMCID: PMC2249765
- DOI: 10.1110/ps.051721006
Mimicking the action of GroEL in molecular dynamics simulations: application to the refinement of protein structures
Abstract
Bacterial chaperonin, GroEL, together with its co-chaperonin, GroES, facilitates the folding of a variety of polypeptides. Experiments suggest that GroEL stimulates protein folding by multiple cycles of binding and release. Misfolded proteins first bind to an exposed hydrophobic surface on GroEL. GroES then encapsulates the substrate and triggers its release into the central cavity of the GroEL/ES complex for folding. In this work, we investigate the possibility to facilitate protein folding in molecular dynamics simulations by mimicking the effects of GroEL/ES namely, repeated binding and release, together with spatial confinement. During the binding stage, the (metastable) partially folded proteins are allowed to attach spontaneously to a hydrophobic surface within the simulation box. This destabilizes the structures, which are then transferred into a spatially confined cavity for folding. The approach has been tested by attempting to refine protein structural models generated using the ROSETTA procedure for ab initio structure prediction. Dramatic improvements in regard to the deviation of protein models from the corresponding experimental structures were observed. The results suggest that the primary effects of the GroEL/ES system can be mimicked in a simple coarse-grained manner and be used to facilitate protein folding in molecular dynamics simulations. Furthermore, the results support the assumption that the spatial confinement in GroEL/ES assists the folding of encapsulated proteins.
Figures
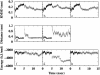


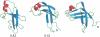


References
-
- Baumketner, A., Jewett, A., and Shea, J.E. 2003. Effects of confinement in chaperonin assisted protein folding: Rate enhancement by decreasing the roughness of the folding energy landscape. J. Mol. Biol. 332: 701–713. - PubMed
-
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., and Hermans, J.1981. Interaction models for water in relation to protein hydration. In Intermolecular forces (ed. B. Pullman), pp. 331–342. Reidel, Dordrecht, The Netherlands.
-
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., Di Nola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81: 3684–3690.
-
- Berendsen, H.J.C., van der Spoel, D., and van Drunen, R. 1995. GRO-MACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91: 43–56.
-
- Betancourt, M.R. and Thirumalai, D. 1999. Exploring the kinetic requirements for enhancement of protein folding rates in the GroEL cavity. J. Mol. Biol. 287: 627–644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

