Site-specific labeling of proteins for single-molecule FRET by combining chemical and enzymatic modification
- PMID: 16452617
- PMCID: PMC2249784
- DOI: 10.1110/ps.051851506
Site-specific labeling of proteins for single-molecule FRET by combining chemical and enzymatic modification
Abstract
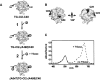
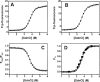
An often limiting factor for studying protein folding by single-molecule fluorescence resonance energy transfer (FRET) is the ability to site-specifically introduce a photostable organic FRET donor (D) and a complementary acceptor (A) into a polypeptide chain. Using alternating-laser excitation and chymotrypsin inhibitor 2 as a model, we show that chemical labeling of a unique cysteine, followed by enzymatic modification of a reactive glutamine in an N-terminally appended substrate sequence recognition tag for transglutaminase (TGase) affords stoichiometrically D-/A-labeled protein suitable for single-molecule FRET experiments. Thermodynamic data indicate that neither the presence of the TGase tag nor D/A labeling perturbs protein stability. As the N terminus in proteins is typically solvent accessible, a TGase tag can (in principle) be appended to any protein of interest by genetic engineering. Two-step chemical/enzymatic labeling may thus represent a simple, low-cost, and widely available strategy for D/A labeling of proteins for FRET-based single-molecule protein folding studies, even for non-protein-experts laboratories.
Figures

Similar articles
-
Site-specific modification of interleukin-2 by the combined use of genetic engineering techniques and transglutaminase.Biochemistry. 1996 Oct 8;35(40):13072-80. doi: 10.1021/bi952616k. Biochemistry. 1996. PMID: 8855943
-
Protein-protein interactions as a tool for site-specific labeling of proteins.Protein Sci. 2005 Aug;14(8):2059-68. doi: 10.1110/ps.051384705. Epub 2005 Jun 29. Protein Sci. 2005. PMID: 15987886 Free PMC article.
-
Transglutaminase-mediated N- and C-terminal fluorescein labeling of a protein can support the native activity of the modified protein.Protein Eng Des Sel. 2004 Feb;17(2):119-26. doi: 10.1093/protein/gzh015. Epub 2004 Jan 12. Protein Eng Des Sel. 2004. PMID: 15047907
-
Site-specific modification and PEGylation of pharmaceutical proteins mediated by transglutaminase.Adv Drug Deliv Rev. 2008 Jan 3;60(1):13-28. doi: 10.1016/j.addr.2007.06.015. Epub 2007 Aug 16. Adv Drug Deliv Rev. 2008. PMID: 17916398 Review.
-
Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics.Biotechnol Lett. 2006 Dec;28(24):1971-82. doi: 10.1007/s10529-006-9193-5. Epub 2006 Oct 5. Biotechnol Lett. 2006. PMID: 17021660 Review.
Cited by
-
Single-Molecule Analysis of Reverse Transcriptase Enzymes.Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a032458. doi: 10.1101/cshperspect.a032458. Cold Spring Harb Perspect Biol. 2019. PMID: 31481455 Free PMC article. Review.
-
Efficient site-specific labeling of proteins via cysteines.Bioconjug Chem. 2008 Mar;19(3):786-91. doi: 10.1021/bc7002499. Epub 2008 Feb 15. Bioconjug Chem. 2008. PMID: 18275130 Free PMC article.
-
Engineering a Prototypic P-type ATPase Listeria monocytogenes Ca(2+)-ATPase 1 for Single-Molecule FRET Studies.Bioconjug Chem. 2016 Sep 21;27(9):2176-87. doi: 10.1021/acs.bioconjchem.6b00387. Epub 2016 Aug 24. Bioconjug Chem. 2016. PMID: 27501274 Free PMC article.
-
Linking time-series of single-molecule experiments with molecular dynamics simulations by machine learning.Elife. 2018 May 3;7:e32668. doi: 10.7554/eLife.32668. Elife. 2018. PMID: 29723137 Free PMC article.
-
A practical guide to single-molecule FRET.Nat Methods. 2008 Jun;5(6):507-16. doi: 10.1038/nmeth.1208. Nat Methods. 2008. PMID: 18511918 Free PMC article. Review.
References
-
- Brasselet, S. and Moerner, W.E. 2000. Fluorescence behaviour of single-molecule pH-sensors. Single Mol. 1: 17–23.
-
- Chin, J.W., Cropp, T.A., Anderson, J.C., Mukherji, M., Zhang, Z., and Schultz, P.G. 2003. An expanded eukaryotic genetic code. Science 301: 964–967. - PubMed
-
- Clegg, R.M. 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211: 353–388. - PubMed
-
- Coussons, P.J., Price, N.C., Kelly, S.M., and Fothergill-Gilmore, L.A. 1992a. The modification of bovine β-casein using transglutaminase purified from guinea pig liver. Biochem. Soc. Trans. 20: 48S. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

