Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus
- PMID: 16452981
- PMCID: PMC1559657
- DOI: 10.1038/nature04487
Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus
Abstract
The critical viral components for packaging DNA, recognizing and binding to host cells, and injecting the condensed DNA into the host are organized at a single vertex of many icosahedral viruses. These component structures do not share icosahedral symmetry and cannot be resolved using a conventional icosahedral averaging method. Here we report the structure of the entire infectious Salmonella bacteriophage epsilon15 (ref. 1) determined from single-particle cryo-electron microscopy, without icosahedral averaging. This structure displays not only the icosahedral shell of 60 hexamers and 11 pentamers, but also the non-icosahedral components at one pentameric vertex. The densities at this vertex can be identified as the 12-subunit portal complex sandwiched between an internal cylindrical core and an external tail hub connecting to six projecting trimeric tailspikes. The viral genome is packed as coaxial coils in at least three outer layers with approximately 90 terminal nucleotides extending through the protein core and the portal complex and poised for injection. The shell protein from icosahedral reconstruction at higher resolution exhibits a similar fold to that of other double-stranded DNA viruses including herpesvirus, suggesting a common ancestor among these diverse viruses. The image reconstruction approach should be applicable to studying other biological nanomachines with components of mixed symmetries.
Figures
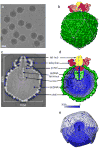


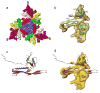
References
-
- McConnell M, Reznick A, Wright A. Studies on the initial interactions of bacteriophage epsilon15 with its host cell, Salmonella anatum. Virology. 1979;94:10–23. - PubMed
-
- Jiang W, et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–5. - PubMed
-
- Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–33. - PubMed
-
- Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

