Sequence variation of the SeM gene of Streptococcus equi allows discrimination of the source of strangles outbreaks
- PMID: 16455902
- PMCID: PMC1392674
- DOI: 10.1128/JCM.44.2.480-486.2006
Sequence variation of the SeM gene of Streptococcus equi allows discrimination of the source of strangles outbreaks
Abstract
Improved understanding of the epidemiology of Streptococcus equi transmission requires sensitive and portable subtyping methods that can rationally discriminate between strains. S. equi is highly homogeneous and cannot be distinguished by multilocus enzyme electrophoretic or multilocus sequence-typing methods that utilize housekeeping genes. However, on sequence analysis of the N-terminal region of the SeM genes of 60 S. equi isolates from 27 strangles outbreaks, we identified 21 DNA codon changes. These resulted in the nonsynonymous substitution of 18 amino acids and allowed the assignment of S. equi strains to 15 distinct subtypes. Our data suggest the presence of multiple epitopes across this region that are subjected to selective immune pressure (nonsynonymous-synonymous substitution rate [d(N)/d(S)] ratio = 3.054), particularly during the establishment of long-term S. equi infection. We further report the application of SeM gene subtyping as a method to investigate potential cases of disease related to administration of a live attenuated S. equi vaccine. SeM gene subtyping successfully differentiated between the vaccine strain and field strains of S. equi responsible for concurrent disease. These results were confirmed by the development and application of a PCR diagnostic test, which identifies the aroA partial gene deletion present in the Equilis StrepE vaccine strain. Although the vaccine strain was found to be responsible for injection site lesions, all seven outbreaks of strangles investigated in recently vaccinated horses were found to be due to concurrent infection with wild-type S. equi and not due to reversion of the vaccine strain.
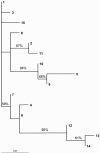
Figures


References
-
- Al-Ghamdi, G. M., V. Kapur, T. R. Ames, J. F. Timoney, D. N. Love, and M. A. Mellencamp. 2000. Use of repetitive sequence-based polymerase chain reaction for molecular epidemiologic analysis of Streptococcus equi subspecies equi. Am. J. Vet. Res. 61:699-705. - PubMed
-
- Chanter, N., N. C. Talbot, J. R. Newton, D. Hewson, and K. Verheyen. 2000. Streptococcus equi with truncated M-proteins isolated from outwardly healthy horses. Microbiology 146:1361-1369. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

