HelF, a putative RNA helicase acts as a nuclear suppressor of RNAi but not antisense mediated gene silencing
- PMID: 16456031
- PMCID: PMC1360742
- DOI: 10.1093/nar/gkj465
HelF, a putative RNA helicase acts as a nuclear suppressor of RNAi but not antisense mediated gene silencing
Abstract
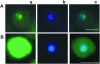
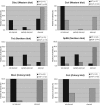
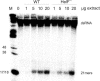
We have identified a putative RNA helicase from Dictyostelium that is closely related to drh-1, the 'dicer-related-helicase' from Caenorhabditis elegans and that also has significant similarity to proteins from vertebrates and plants. Green fluorescent protein (GFP)-tagged HelF protein was localized in speckles in the nucleus. Disruption of the helF gene resulted in a mutant morphology in late development. When transformed with RNAi constructs, HelF- cells displayed enhanced RNA interference on four tested genes. One gene that could not be knocked-down in the wild-type background was efficiently silenced in the mutant. Furthermore, the efficiency of silencing in the wild-type was dramatically improved when helF was disrupted in a secondary transformation. Silencing efficiency depended on transcription levels of hairpin RNA and the threshold was dramatically reduced in HelF- cells. However, the amount of siRNA did not depend on hairpin transcription. HelF is thus a natural nuclear suppressor of RNA interference. In contrast, no improvement of gene silencing was observed when mutant cells were challenged with corresponding antisense constructs. This indicates that RNAi and antisense have distinct requirements even though they may share parts of their pathways.
Figures




Similar articles
-
RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs.Science. 2002 Jan 25;295(5555):694-7. doi: 10.1126/science.1067534. Science. 2002. PMID: 11809977
-
RNA interference and antisense-mediated gene silencing in Dictyostelium.Methods Mol Biol. 2006;346:211-26. doi: 10.1385/1-59745-144-4:211. Methods Mol Biol. 2006. PMID: 16957293
-
The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans.Cell. 2002 Jun 28;109(7):861-71. doi: 10.1016/s0092-8674(02)00793-6. Cell. 2002. PMID: 12110183
-
Antisense-RNA regulation and RNA interference.Biochim Biophys Acta. 2002 May 3;1575(1-3):15-25. doi: 10.1016/s0167-4781(02)00280-4. Biochim Biophys Acta. 2002. PMID: 12020814 Review.
-
Structural and functional modules in RNA interference.Curr Opin Struct Biol. 2009 Jun;19(3):286-93. doi: 10.1016/j.sbi.2009.04.006. Epub 2009 May 26. Curr Opin Struct Biol. 2009. PMID: 19477631 Free PMC article. Review.
Cited by
-
Evidence that noncoding RNA dutA is a multicopy suppressor of Dictyostelium discoideum STAT protein Dd-STATa.Eukaryot Cell. 2007 Jun;6(6):1030-40. doi: 10.1128/EC.00035-07. Epub 2007 Apr 13. Eukaryot Cell. 2007. PMID: 17435008 Free PMC article.
-
MicroRNAs in Amoebozoa: deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs.RNA. 2012 Oct;18(10):1771-82. doi: 10.1261/rna.033175.112. Epub 2012 Aug 8. RNA. 2012. PMID: 22875808 Free PMC article.
-
Analysis of chikungunya virus proteins reveals that non-structural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity.Sci Rep. 2016 Nov 30;6:38065. doi: 10.1038/srep38065. Sci Rep. 2016. PMID: 27901124 Free PMC article.
-
Dictyostelium finds new roles to model.Genetics. 2010 Jul;185(3):717-26. doi: 10.1534/genetics.110.119297. Genetics. 2010. PMID: 20660652 Free PMC article. Review.
-
ABCE1 is a highly conserved RNA silencing suppressor.PLoS One. 2015 Feb 6;10(2):e0116702. doi: 10.1371/journal.pone.0116702. eCollection 2015. PLoS One. 2015. PMID: 25659154 Free PMC article.
References
-
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. - PubMed
-
- Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. - PubMed
-
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

