Engineering a rare-cutting restriction enzyme: genetic screening and selection of NotI variants
- PMID: 16456032
- PMCID: PMC1360745
- DOI: 10.1093/nar/gkj483
Engineering a rare-cutting restriction enzyme: genetic screening and selection of NotI variants
Abstract
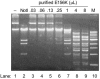
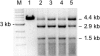
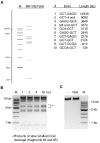
Restriction endonucleases (REases) with 8-base specificity are rare specimens in nature. NotI from Nocardia otitidis-caviarum (recognition sequence 5'-GCGGCCGC-3') has been cloned, thus allowing for mutagenesis and screening for enzymes with altered 8-base recognition and cleavage activity. Variants possessing altered specificity have been isolated by the application of two genetic methods. In step 1, variant E156K was isolated by its ability to induce DNA-damage in an indicator strain expressing M.EagI (to protect 5'-NCGGCCGN-3' sites). In step 2, the E156K allele was mutagenized with the objective of increasing enzyme activity towards the alternative substrate site: 5'-GCTGCCGC-3'. In this procedure, clones of interest were selected by their ability to eliminate a conditionally toxic substrate vector and induce the SOS response. Thus, specific DNA cleavage was linked to cell survival. The secondary substitutions M91V, F157C and V348M were each found to have a positive effect on specific activity when paired with E156K. For example, variant M91V/E156K cleaves 5'-GCTGCCGC-3' with a specific activity of 8.2 x 10(4) U/mg, a 32-fold increase over variant E156K. A comprehensive analysis indicates that the cleavage specificity of M91V/E156K is relaxed to a small set of 8 bp substrates while retaining activity towards the NotI sequence.
Figures



References
-
- Heitman J., Model P. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene. 1991;103:1–9. - PubMed
-
- Samuelson J.C., Xu S.Y. Directed evolution of restriction endonuclease BstYI to achieve increased substrate specificity. J. Mol. Biol. 2002;319:673–683. - PubMed
-
- Townson S.A., Samuelson J.C., Xu S.Y., Aggarwal A.K. Implications for switching restriction enzyme specificities from the structure of BstYI bound to a BglII DNA sequence. Structure (Camb) 2005;13:791–801. - PubMed
-
- Zhu Z., Zhou J., Friedman A.M., Xu S.Y. Isolation of BsoBI restriction endonuclease variants with altered substrate specificity. J. Mol. Biol. 2003;330:359–372. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

