Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells
- PMID: 16456034
- PMCID: PMC1360746
- DOI: 10.1093/nar/gkj495
Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells
Abstract
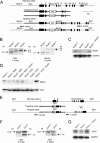
The Mus81-Eme1 complex is a structure-specific endonuclease that preferentially cleaves nicked Holliday junctions, 3'-flap structures and aberrant replication fork structures. Mus81-/- mice have been shown to exhibit spontaneous chromosomal aberrations and, in one of two models, a predisposition to cancers. The molecular mechanisms underlying its role in chromosome integrity, however, are largely unknown. To clarify the role of Mus81 in human cells, we deleted the gene in the human colon cancer cell line HCT116 by gene targeting. Here we demonstrate that Mus81 confers resistance to DNA crosslinking agents and slight resistance to other DNA-damaging agents. Mus81 deficiency spontaneously promotes chromosome damage such as breaks and activates the intra-S-phase checkpoint through the ATM-Chk1/Chk2 pathways. Furthermore, Mus81 deficiency activates the G2/M checkpoint through the ATM-Chk2 pathway and promotes DNA rereplication. Increased rereplication is reversed by the ectopic expression of Cdk1. Haploinsufficiency of Mus81 or Eme1 also causes similar phenotypes. These findings suggest that a complex network of the checkpoint pathways that respond to DNA double-strand breaks may participate in some of the phenotypes associated with Mus81 or Eme1 deficiency.
Figures




Similar articles
-
Involvement of mammalian Mus81 in genome integrity and tumor suppression.Science. 2004 Jun 18;304(5678):1822-6. doi: 10.1126/science.1094557. Science. 2004. PMID: 15205536
-
Functional interplay of p53 and Mus81 in DNA damage responses and cancer.Cancer Res. 2007 Sep 15;67(18):8527-35. doi: 10.1158/0008-5472.CAN-07-1161. Cancer Res. 2007. PMID: 17875692
-
Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks.DNA Repair (Amst). 2007 Jul 1;6(7):1004-17. doi: 10.1016/j.dnarep.2007.02.019. Epub 2007 Apr 3. DNA Repair (Amst). 2007. PMID: 17409028 Review.
-
Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation.PLoS One. 2011;6(8):e23517. doi: 10.1371/journal.pone.0023517. Epub 2011 Aug 17. PLoS One. 2011. PMID: 21858151 Free PMC article.
-
Structure-specific endonucleases: guardians of fragile site stability.Trends Cell Biol. 2014 May;24(5):321-7. doi: 10.1016/j.tcb.2013.11.007. Epub 2013 Dec 19. Trends Cell Biol. 2014. PMID: 24361091 Review.
Cited by
-
Effect of EME1 exon variant Ile350Thr on risk and early onset of breast cancer in southern Chinese women.J Biomed Res. 2013 May;27(3):193-201. doi: 10.7555/JBR.27.20130013. Epub 2013 Apr 15. J Biomed Res. 2013. PMID: 23720674 Free PMC article.
-
Cooperativity of Mus81.Mms4 with Rad54 in the resolution of recombination and replication intermediates.J Biol Chem. 2009 Mar 20;284(12):7733-45. doi: 10.1074/jbc.M806192200. Epub 2009 Jan 7. J Biol Chem. 2009. Retraction in: J Biol Chem. 2024 Oct;300(10):107747. doi: 10.1016/j.jbc.2024.107747. PMID: 19129197 Free PMC article. Retracted.
-
Haploinsufficiency of DNA Damage Response Genes and their Potential Influence in Human Genomic Disorders.Curr Genomics. 2008 May;9(3):137-46. doi: 10.2174/138920208784340795. Curr Genomics. 2008. PMID: 19440510 Free PMC article.
-
Radiosensitisation of human colorectal cancer cells by ruthenium(II) arene anticancer complexes.Sci Rep. 2016 Feb 12;6:20596. doi: 10.1038/srep20596. Sci Rep. 2016. PMID: 26867983 Free PMC article.
-
The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage.Nat Commun. 2015 Apr 16;6:6746. doi: 10.1038/ncomms7746. Nat Commun. 2015. PMID: 25879486 Free PMC article.
References
-
- Branzei D., Foiani M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005;17:568–575. - PubMed
-
- Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. - PubMed
-
- Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. - PubMed
-
- Interthal H., Heyer H.-D. MUS81 encodes a novel Helix-hairpin-Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

