A suboptimal 5' splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication
- PMID: 16457729
- PMCID: PMC1403798
- DOI: 10.1186/1742-4690-3-10
A suboptimal 5' splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication
Abstract
Background: Inefficient alternative splicing of the human immunodeficiency virus type 1(HIV-1) primary RNA transcript results in greater than half of all viral mRNA remaining unspliced. Regulation of HIV-1 alternative splicing occurs through the presence of suboptimal viral 5' and 3' splice sites (5' and 3'ss), which are positively regulated by exonic splicing enhancers (ESE) and negatively regulated by exonic splicing silencers (ESS) and intronic splicing silencers (ISS). We previously showed that splicing at HIV-1 3'ss A2 is repressed by ESSV and enhanced by the downstream 5'ss D3 signal. Disruption of ESSV results in increased vpr mRNA accumulation and exon 3 inclusion, decreased accumulation of unspliced viral mRNA, and decreased virus production.
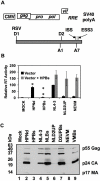
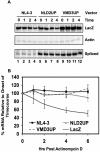
Results: Here we show that optimization of the 5'ss D2 signal results in increased splicing at the upstream 3'ss A1, increased inclusion of exon 2 into viral mRNA, decreased accumulation of unspliced viral mRNA, and decreased virus production. Virus production from the 5'ss D2 and ESSV mutants was rescued by transient expression of HIV-1 Gag and Pol. We further show that the increased inclusion of either exon 2 or 3 does not significantly affect the stability of viral mRNA but does result in an increase and decrease, respectively, in HIV-1 mRNA levels. The changes in viral mRNA levels directly correlate with changes in tat mRNA levels observed upon increased inclusion of exon 2 or 3.
Conclusion: These results demonstrate that splicing at HIV-1 3'ss A1 is regulated by the strength of the downstream 5'ss signal and that suboptimal splicing at 3'ss A1 is necessary for virus replication. Furthermore, the replication defective phenotype resulting from increased splicing at 3'ss A1 is similar to the phenotype observed upon increased splicing at 3'ss A2. Further examination of the role of 5'ss D2 and D3 in the alternative splicing of 3'ss A1 and A2, respectively, is necessary to delineate a role for non-coding exon inclusion in HIV-1 replication.
Figures


Similar articles
-
Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression.J Virol. 2008 Apr;82(8):3921-31. doi: 10.1128/JVI.01558-07. Epub 2008 Feb 13. J Virol. 2008. PMID: 18272582 Free PMC article.
-
An exonic splicing silencer downstream of the 3' splice site A2 is required for efficient human immunodeficiency virus type 1 replication.J Virol. 2005 Aug;79(16):10478-86. doi: 10.1128/JVI.79.16.10478-10486.2005. J Virol. 2005. PMID: 16051840 Free PMC article.
-
Tra2-mediated recognition of HIV-1 5' splice site D3 as a key factor in the processing of vpr mRNA.J Virol. 2013 Mar;87(5):2721-34. doi: 10.1128/JVI.02756-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255807 Free PMC article.
-
Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication.Adv Virus Res. 2009;74:1-40. doi: 10.1016/S0065-3527(09)74001-1. Adv Virus Res. 2009. PMID: 19698894 Review.
-
Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing.Curr HIV Res. 2006 Jan;4(1):43-55. doi: 10.2174/157016206775197655. Curr HIV Res. 2006. PMID: 16454710 Review.
Cited by
-
Differential effects of hnRNP D/AUF1 isoforms on HIV-1 gene expression.Nucleic Acids Res. 2012 Apr;40(8):3663-75. doi: 10.1093/nar/gkr1238. Epub 2011 Dec 19. Nucleic Acids Res. 2012. PMID: 22187150 Free PMC article.
-
The strength of the HIV-1 3' splice sites affects Rev function.Retrovirology. 2006 Dec 4;3:89. doi: 10.1186/1742-4690-3-89. Retrovirology. 2006. PMID: 17144911 Free PMC article.
-
Gag-processing defect of human immunodeficiency virus type 1 integrase E246 and G247 mutants is caused by activation of an overlapping 5' splice site.J Virol. 2008 Feb;82(3):1600-4. doi: 10.1128/JVI.02295-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032510 Free PMC article.
-
Regulation of Vif mRNA splicing by human immunodeficiency virus type 1 requires 5' splice site D2 and an exonic splicing enhancer to counteract cellular restriction factor APOBEC3G.J Virol. 2009 Jun;83(12):6067-78. doi: 10.1128/JVI.02231-08. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357165 Free PMC article.
-
Replication of human immunodeficiency virus type 1 from entry to exit.Int J Hematol. 2006 Jul;84(1):23-30. doi: 10.1532/IJH97.06112. Int J Hematol. 2006. PMID: 16867898 Review.
References
-
- Neumann M, Harrison J, Saltarelli M, Hadziyannis E, Erfle V, Felber BK, Pavlakis GN. Splicing variability in HIV type 1 revealed by quantitative RNA polymerase chain reaction. AIDS Res Hum Retroviruses. 1994;10:1531–1542. - PubMed
-
- Si Z, Amendt BA, Stoltzfus CM. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3' splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

