The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control
- PMID: 16461653
- PMCID: PMC1392936
- DOI: 10.1128/AEM.72.2.1086-1095.2006
The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control
Abstract
Polaromonas naphthalenivorans CJ2, found to be responsible for the degradation of naphthalene in situ at a coal tar waste-contaminated site (C.-O. Jeon et al., Proc. Natl. Acad. Sci. USA 100:13591-13596, 2003), is able to grow on mineral salts agar media with naphthalene as the sole carbon source. Beginning from a 484-bp nagAc-like region, we used a genome walking strategy to sequence genes encoding the entire naphthalene degradation pathway andadditional flanking regions. We found that the naphthalene catabolic genes in P. naphthalenivorans CJ2 were divided into one large and one small gene cluster, separated by an unknown distance. The large gene cluster (nagRAaGHAbAcAdBFCQEDJI'ORF1tnpA) is bounded by a LysR-type regulator (nagR). The small cluster (nagR2ORF2I"KL) is bounded by a MarR-type regulator (nagR2). The catabolic genes of P. naphthalenivorans CJ2 were homologous to many of those of Ralstonia U2, which uses the gentisate pathway to convert naphthalene to central metabolites. However, three open reading frames (nagY, nagM, and nagN), present in Ralstonia U2, were absent. Also, P. naphthalenivorans carries two copies of gentisate dioxygenase (nagI) with 77.4% DNA sequence identity to one another and 82% amino acid identity to their homologue in Ralstonia sp. strain U2. Investigation of the operons using reverse transcription PCR showed that each cluster was controlled independently by its respective promoter. Insertional inactivation and lacZ reporter assays showed that nagR2 is a negative regulator and that expression of the small cluster is not induced by naphthalene, salicylate, or gentisate. Association of two putative Azoarcus-related transposases with the large cluster and one Azoarcus-related putative salicylate 5-hydroxylase gene (ORF2) in the small cluster suggests that mobile genetic elements were likely involved in creating the novel arrangement of catabolic and regulatory genes in P. naphthalenivorans.
Figures
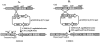





References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Assinder, S. J., and P. A. Williams. 1988. Comparison of the meta-pathway operons on NAH plasmid pWW60-22 and TOL plasmid pWW53-4 and its evolutionary significance. J. Gen. Microbiol. 234:2769-2778. - PubMed
-
- Bosch, R., E. Garcia-Valdes, and E. R. B. Moore. 1999. Genetic characterization and evolutionary implication of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149-157. - PubMed
-
- Bosch, R., E. Garcia-Valdes, and E. R. B. Moore. 2000. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245:65-74. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

