Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752
- PMID: 16461907
- PMCID: PMC1413705
- DOI: 10.1073/pnas.0508776103
Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752
Erratum in
-
Correction for Smolen et al., Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2410435121. doi: 10.1073/pnas.2410435121. Epub 2024 Jun 13. Proc Natl Acad Sci U S A. 2024. PMID: 38870064 Free PMC article. No abstract available.
Abstract
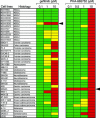
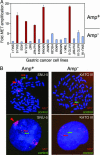
The success of molecular targeted therapy in cancer may depend on the selection of appropriate tumor types whose survival depends on the drug target, so-called "oncogene addiction." Preclinical approaches to defining drug-responsive subsets are needed if initial clinical trials are to be directed at the most susceptible patient population. Here, we show that gastric cancer cells with high-level stable chromosomal amplification of the growth factor receptor MET are extraordinarily susceptible to the selective inhibitor PHA-665752. Although MET activation has primarily been linked with tumor cell migration and invasiveness, the amplified wild-type MET in these cells is constitutively activated, and its continued signaling is required for cell survival. Treatment with PHA-665752 triggers massive apoptosis in 5 of 5 gastric cancer cell lines with MET amplification but in 0 of 12 without increased gene copy numbers (P = 0.00016). MET amplification may thus identify a subset of epithelial cancers that are uniquely sensitive to disruption of this pathway and define a patient group that is appropriate for clinical trials of targeted therapy using MET inhibitors.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Weinstein I. B. Science. 2002;297:63–64. - PubMed
-
- Apperley J. F., Gardembas M., Melo J. V., Russell-Jones R., Bain B. J., Baxter E. J., Chase A., Chessells J. M., Colombat M., Dearden C. E., et al. N. Engl. J. Med. 2002;347:481–487. - PubMed
-
- Druker B. J., Talpaz M., Resta D. J., Peng B., Buchdunger E., Ford J. M., Lydon N. B., Kantarjian H., Capdeville R., Ohno-Jones, et al. N. Engl. J. Med. 2001;344:1031–1037. - PubMed
-
- Heinrich M. C., Corless C. L., Demetri G. D., Blanke C. D., von Mehren M., Joensuu H., McGreevey L. S., Chen C. J., Van den Abbeele A. D., Druker B. J., et al. J. Clin. Oncol. 2003;21:4342–4349. - PubMed
-
- Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., et al. N. Engl. J. Med. 2004;350:2129–2139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

