Reciprocal actions of REST and a microRNA promote neuronal identity
- PMID: 16461918
- PMCID: PMC1413753
- DOI: 10.1073/pnas.0511041103
Reciprocal actions of REST and a microRNA promote neuronal identity
Abstract
MicroRNAs (miRNAs) are implicated in both tissue differentiation and maintenance of tissue identity. In most cases, however, the mechanisms underlying their regulation are not known. One brain-specific miRNA, miR-124a, decreases the levels of hundreds of nonneuronal transcripts, such that its introduction into HeLa cells promotes a neuronal-like mRNA profile. The transcriptional repressor, RE1 silencing transcription factor (REST), has a reciprocal activity, inhibiting the expression of neuronal genes in nonneuronal cells. Here, we show that REST regulates the expression of a family of miRNAs, including brain-specific miR-124a. In nonneuronal cells and neural progenitors, REST inhibits miR-124a expression, allowing the persistence of nonneuronal transcripts. As progenitors differentiate into mature neurons, REST leaves miR-124a gene loci, and nonneuronal transcripts are degraded selectively. Thus, the combined transcriptional and posttranscriptional consequences of REST action maximize the contrast between neuronal and nonneuronal cell phenotypes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
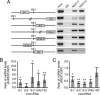

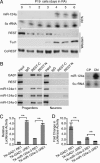
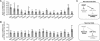
References
-
- Edlund T., Jessell T. M. Cell. 1999;96:211–224. - PubMed
-
- Munoz-San Juan I., Brivanlou A. H. Nat. Rev. Neurosci. 2002;3:271–280. - PubMed
-
- Chong J. A., Tapia-Ramirez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., Mandel G. Cell. 1995;80:949–957. - PubMed
-
- Schoenherr C. J., Anderson D. J. Science. 1995;267:1360–1363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

